Unraveling the Genetic Basis of Sorghum Plant Color: Insights into Phenotypic Traits and Fungal Resistance
In sorghum, plant color is a key trait determined by the P and Q genes, with different genotypes resulting in purple (P_Q_), red (P_ qq), or tan (pp Q_ and pp qq) coloration. Beyond aesthetics, plant color is linked to various phenotypes and consumer preferences. For instance, white sorghum grains from tan plants are preferred for both human and animal consumption due to lower tannin content, while purple/red plants have higher levels of total phenols. However, tan plants exhibit lower susceptibility to head blight caused by Fusarium moniliforme, a major fungal pathogen of cereals. Flavonoids such as luteolin, found in higher levels in tan plants, act as phytoalexins against pathogens like Colletotrichum sublineolum. The underlying genetics of plant color involve genes like Sobic.006G226800 (Sb06g029550), which encodes a protein with flavanone 4-reductase activity.
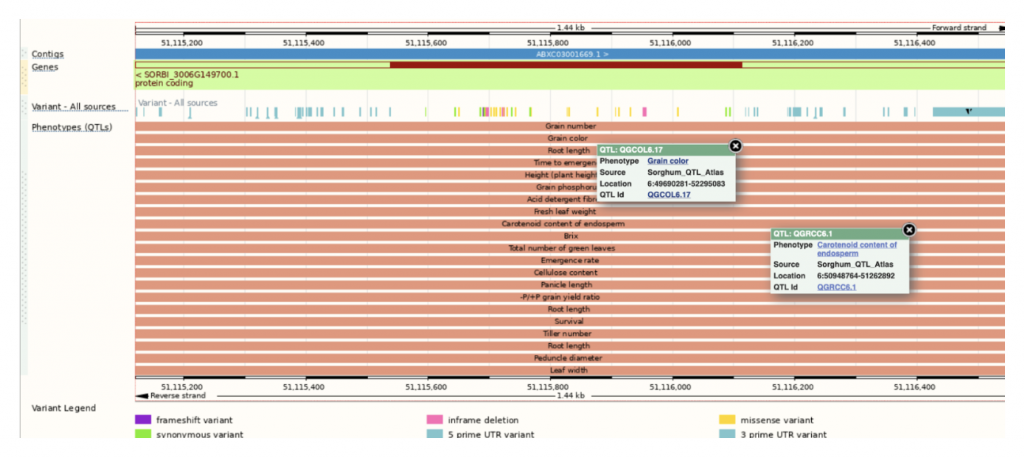
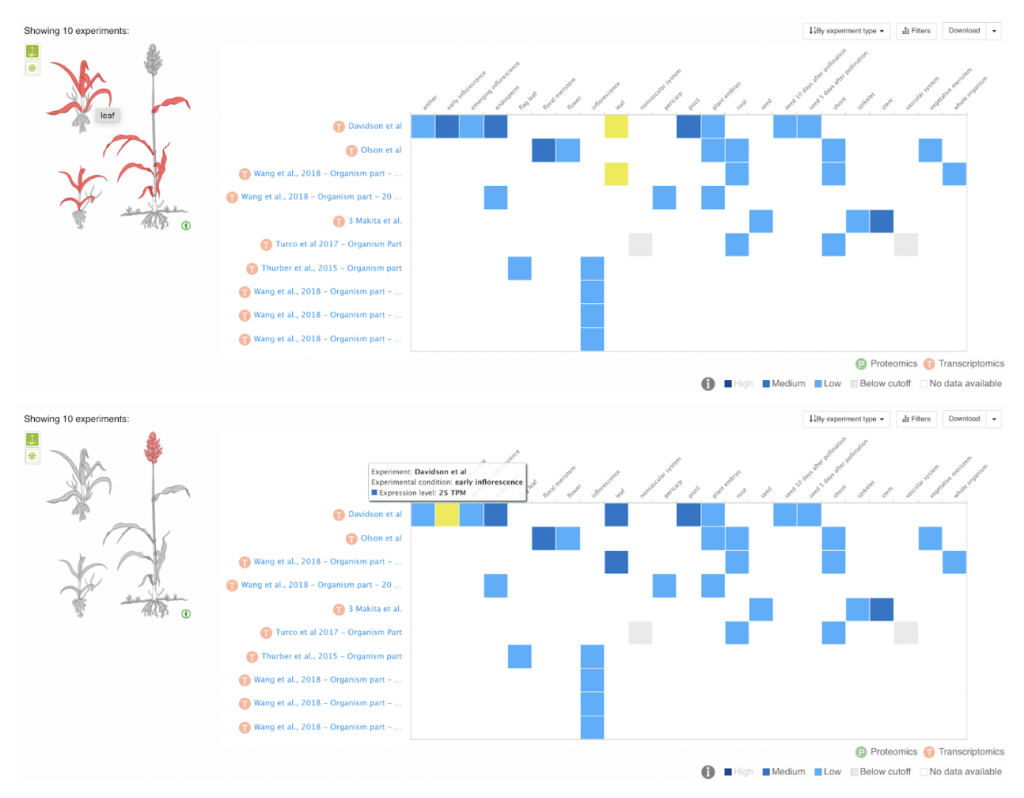
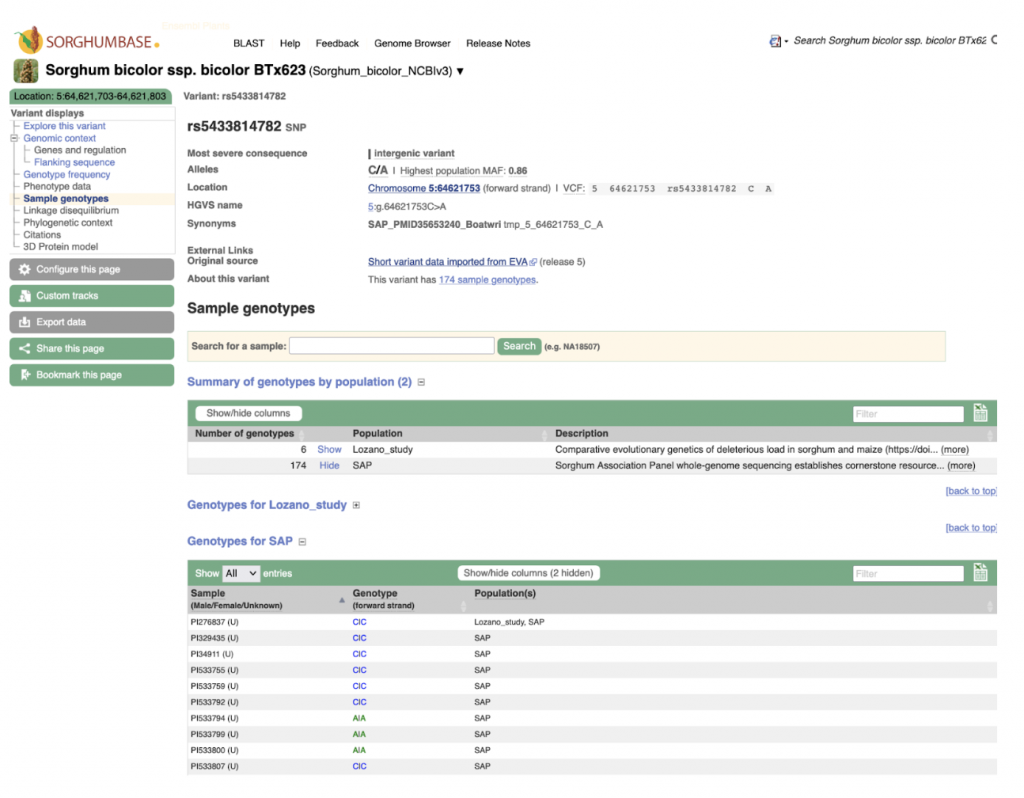
In a recent study, researchers from Anhui Science and Technology University, Anhui Province International Joint Research Center of Forage Bio-breeding and University of Louisiana at Lafayette assessed the plant color of the sorghum mini core collection by scoring the leaf sheath and leaf color at maturity as tan, red, or purple across three different testing environments in China. They conducted association mapping using over 6 million SNP markers and identified candidate genes in eight loci that are strongly linked to plant color. Notably, the qPC6 locus on chromosome 6 was found to be distinct from previously identified QTLs, suggesting multiple genes may control plant color. Candidate genes linked to these loci, such as Sobic.006G149700, a senescence regulator, showed tissue-specific expression patterns, indicating their potential roles in plant color determination. Additionally, they identified a candidate gene, Sobic.005G165700, associated with fungal resistance, highlighting the multifaceted nature of plant color development and its interaction with environmental factors like pathogen attack. The authors found a loose correlation between the degree of linkage and tissue/organ expression of the underlying genes possibly related to the plant color phenotype. Furthermore, allele analysis indicated that none of the linked SNPs can differentiate between red and purple accessions whereas all linked SNPs can differentiate tan from red/purple accessions. These findings provide some insights into the genetics underlying sorghum plant color and offer opportunities to test candidate genes for targeted breeding efforts to enhance desired traits.
SorghumBase examples:
Reference:
Wang L, Tu W, Jin P, Liu Y, Du J, Zheng J, Wang YH, Li J. Genome-wide association study of plant color in Sorghum bicolor. Front Plant Sci. 2024 Apr 10;15:1320844. PMID: 38660439. doi: 10.3389/fpls.2024.1320844. Read more

