Unveiling Molecular Mechanisms of Sorghum Resistance to Sugarcane Aphids Through Transcriptomic Analysis
Scientists from Oklahoma State University and USDA-ARS investigated the molecular mechanisms of sorghum’s defense against sugarcane aphids through an integrated transcriptomic and pathway analysis. The research involved examining the responses of resistant (Tx2783) and susceptible (BTx623) sorghum genotypes to aphid infestation over various time points (1, 3, 6, 9, and 12 days post-infestation, dpi). The team, led by Yinghua Huang, utilized RNA-seq to identify differentially expressed genes (DEGs) between the genotypes. The analysis revealed a significant number of DEGs, with the resistant genotype showing a gradual increase in upregulated DEGs and a decrease in downregulated DEGs over time. In contrast, the susceptible genotype had a higher number of DEGs overall, indicating a more pronounced response to aphid infestation.
The study also included a principal component analysis (PCA) to explore the statistical correlations between the genotypes and time points, which effectively separated the resistant and susceptible genotypes. The PCA indicated that the first principal component, accounting for 34.44% of the variance, distinguished between the genotypes’ differential responses, while the second component, accounting for 18.32% of the variance, showed differences between the time points.
Gene ontology and KEGG pathway analyses were conducted to understand the functions of the DEGs. The findings highlighted the role of various genes and pathways in the sorghum’s defense mechanisms, including those related to signal perception, transduction, and defense response. The study also noted the importance of phytohormones like salicylic acid (SA) and jasmonic acid (JA) in reducing aphid populations and damage in the susceptible genotype.
In addition it discusses the role of TFs in regulating gene expression during biotic stress, specifically in the context of sorghum’s response to sugarcane aphid infestation. Several TF families were highlighted, including WRKY, MYB, HD-Zip, and EREBP, which are differentially expressed in both resistant and susceptible sorghum genotypes. The WRKY TFs are noted for their involvement in redox homeostasis, salicylic acid (SA) signaling, and the cross-communication between ethylene (ET) and jasmonic acid (JA), which are crucial for defense against biotic stresses. The study found that WRKY22 and WRKY33 genes were induced in both genotypes across all time points, suggesting their consistent role in the plant’s defense response. MYB TFs are mentioned for their role in activating hypersensitive cell death during pathogen attack and insect feeding, through the regulation of long-chain fatty acid synthesis. The document states that four MYB TFs were highly upregulated in the resistant genotype, indicating their potential importance in the resistant response to aphid infestation. The HD-Zip TFs are unique to the plant kingdom and have been implicated in the regulation of abscisic acid (ABA) homeostasis and signaling, as well as in protecting plants from pathogens and abiotic stresses. The EREBP TFs are involved in hormone signal transduction pathways, including ET, ABA, cytokinin, and JA, and were also identified as differentially expressed in sorghum genotypes during aphid infestation.
Curated genes in the paper: The gene identifiers in Table1 correspond to differentially expressed genes (DEGs) in sorghum genotypes that were studied for their response to sugarcane aphid infestation. The genes are involved in various biological processes and pathways related to the plant’s defense mechanisms.
Table1:
| Gene ID | Description |
| SORBI_3006G183200 | This gene was expressed nine-fold in the resistant genotype across all time points. |
| SORBI_3001G397900 | One of the four MYB genes that were highly upregulated in the resistant genotype but downregulated in the susceptible genotype. |
| SORBI_3008G055700 | Another of the four MYB genes with the same expression pattern as SORBI_3001G397900. |
| SORBI_3004G216900 | The third of the four MYB genes with the same expression pattern as the previous two. |
| SORBI_3002G423300 | One of the two MYB genes that were upregulated in both genotypes. |
| SORBI_3002G337800 | The second of the two MYB genes that were upregulated in both genotypes. |
| SORBI_3001G473900 | One of the two EREBP TF genes that were upregulated more than six-fold across the time points in the two genotypes. |
| SORBI_3006G168000 | The second of the two EREBP TF genes with the same expression pattern as SORBI_3001G473900. |
| SORBI_3005G192100 | Belonging to the RPM1/RPS3 gene family, this gene was upregulated more than four-fold in both genotypes across all time points. It is a putative ortholog to stripe rust resistance protein Yr10 in wheat. |
| SORBI_3001G107200 | The sorghum housekeeping gene, a-Tubulin, used as the internal control in the RT-qPCR analysis. |
A list of all genes mentioned in the study can be found in SorghumBase.
Sorghumbase examples
An example gene SORBI_3005G137000 mentioned in the study is one of the seven chalcone synthase (CHS) genes that were found to be upregulated in the resistant sorghum genotype Tx2783 when exposed to sugarcane aphids. CHS is a key enzyme in the flavonoid biosynthesis pathway, which is part of the larger phenylpropanoid pathway. This pathway is responsible for the synthesis of a wide range of secondary metabolites, including flavonoids, which are important for plant defense against biotic stresses such as insect herbivory. The upregulation of SORBI_3005G137000 and other CHS genes in the resistant genotype suggests that the plant is enhancing the production of flavonoids in response to aphid infestation.
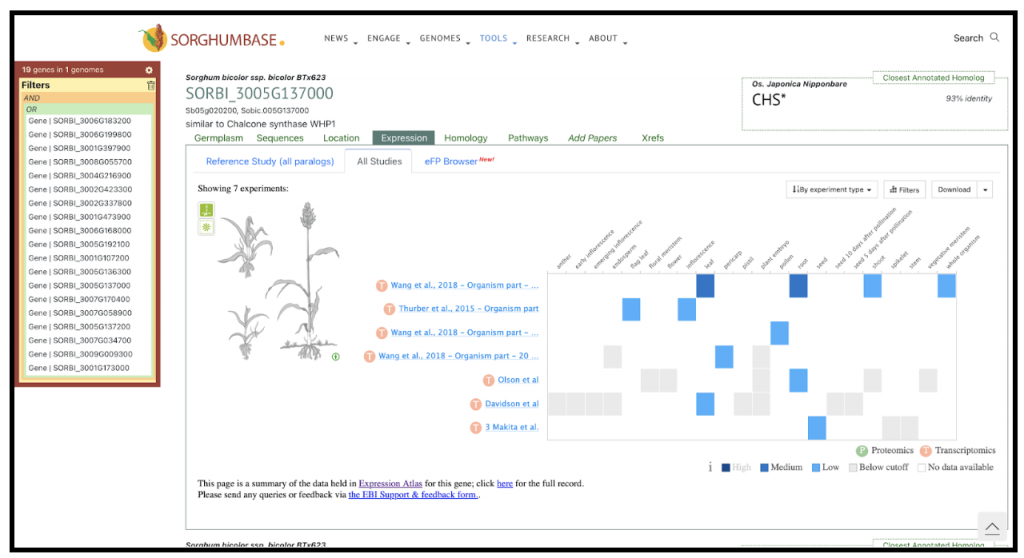
Chalcone synthase catalyzes the first committed step in the flavonoid pathway, converting 4-coumaroyl-CoA and three molecules of malonyl-CoA into naringenin chalcone, which is then isomerized to form the flavanone naringenin. This reaction is the entry point into the flavonoid pathway, and the subsequent enzymatic steps lead to the production of various flavonoid compounds with diverse biological activities.Flavonoids can act as feeding deterrents, toxins, or as signaling molecules that attract natural enemies of the herbivores. They can also contribute to the plant’s defense by reinforcing cell walls or by acting as antioxidants to mitigate the oxidative stress caused by the aphid’s feeding activity.
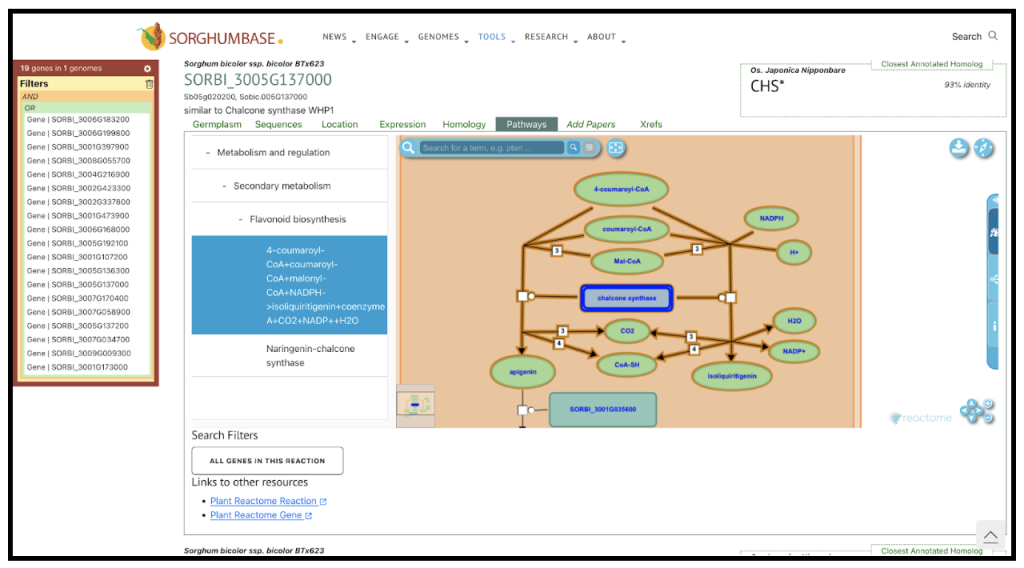
In summary, SORBI_3005G137000 is a gene that encodes for the chalcone synthase enzyme, which plays a crucial role in the flavonoid biosynthesis pathway. Its upregulation in the resistant sorghum genotype indicates its involvement in the plant’s defense mechanism against sugarcane aphids, likely through the increased production of flavonoids that can deter or harm the aphids.
Reference:
Shrestha K, Huang J, Yan L, Doust AN, Huang Y. Integrated transcriptomic and pathway analyses of sorghum plants revealed the molecular mechanisms of host defense against aphids. Front Plant Sci. 2024 Jun 6;15:1324085. PMID: 38903420. doi: 10.3389/fpls.2024.1324085. Read more
Related Project Websites:
- Andrew N. Doust lab at Oklahoma State University: https://cas.okstate.edu/plant_biology/about_us/dr_dousts_lab/

