Unveiling Conservation in Plant Extracellular Vesicle Proteomes: A Sorghum and Arabidopsis Study
Plant extracellular vesicles (EVs) are pivotal in intercellular communication and defense responses. A recent study by scientists from the University of Delaware, Indiana University and Brookhaven National Laboratory delved into the EV proteomes of Sorghum bicolor and Arabidopsis thaliana, revealing intriguing insights into the partial conservation between these plant species. The isolation of EVs from sorghum leaves showcased a heterogeneous population with distinct lipid bilayers and content. Advanced techniques, including nanoparticle tracking analysis and cryo-electron tomography, contributed to the characterization of sorghum EVs. Proteomic analysis identified 437 proteins enriched in sorghum EVs, with a significant overlap observed in the EV proteome of Arabidopsis. This suggests a partial conservation in both function and contents between the monocot sorghum and the distantly related eudicot Arabidopsis.
Despite the extensive research on mammalian EVs, the field of plant EVs faces challenges, necessitating further exploration into fundamental aspects such as biogenesis, morphology, cargo packaging, and interkingdom movement. The study addressed these challenges by adapting an Arabidopsis EV isolation protocol for sorghum. Optimization techniques, including leaf tip snipping and efficient buffer retention, were introduced to enhance EV capture during vacuum infiltration. Additionally, homologs of the marker proteins like PENETRATION1 (PEN1, AT3G11820.1), PATELLIN 1 (PATL1, AT1G72150.1), the ABC transporter PENETRATION3 (PEN3, AT1G59870.1), and TETRASPANIN8 (TET8, AT2G23810.1), as well as the proposed RNA-binding annexin proteins ANN1 (AT1G35720.1) and ANN2 (AT5G65020.1) were identified, laying the groundwork for future investigations in sorghum and other monocots. Biological function GO term enrichment of sorghum orthologs suggested a core of proteins enriched for vesicle docking/fusion, transport across membranes, exocytosis, carbohydrate metabolism, and translation. Cryo-electron tomography unveiled the diverse population of sorghum EVs, while attempts to stain them with fluorescent dyes provided crucial insights into the limitations of commonly used lipophilic dyes. Overall, this research contributes to the evolving understanding of plant EVs, paving the way for deeper exploration into the intricate world of intercellular communication and defense responses in plants.
The goal of this manuscript was to provide a robust extracellular vesicle isolation protocol for sorghum. With this in hand, we were able to show that 60% of the sorghum EV proteins identified have orthologs in the Arabidopsis EV proteome. We hope that our isolation method and characterization by proteomics and microscopy can be used as a guide for other monocot EV research and further our understanding of the key proteins and functions of EVs in plants. – Chaya
SorghumBase examples:
AT1G72150 PATL1 was picked up as an example to get sorghum orthologs to explore SorghumBase below:

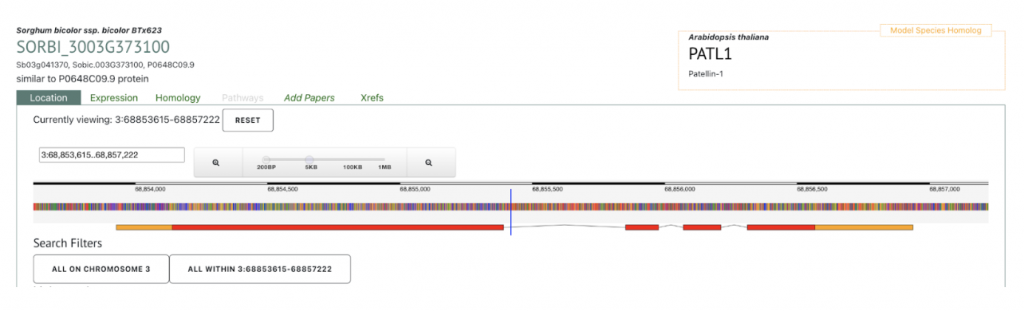

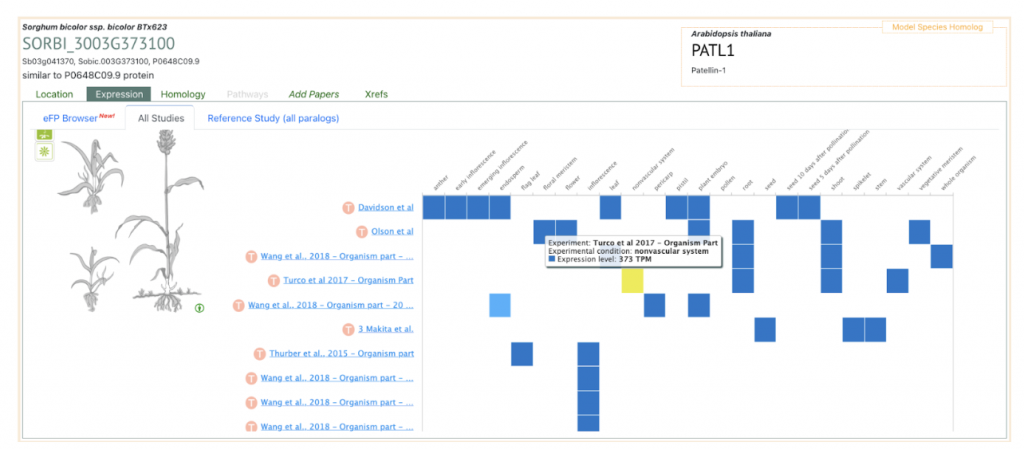
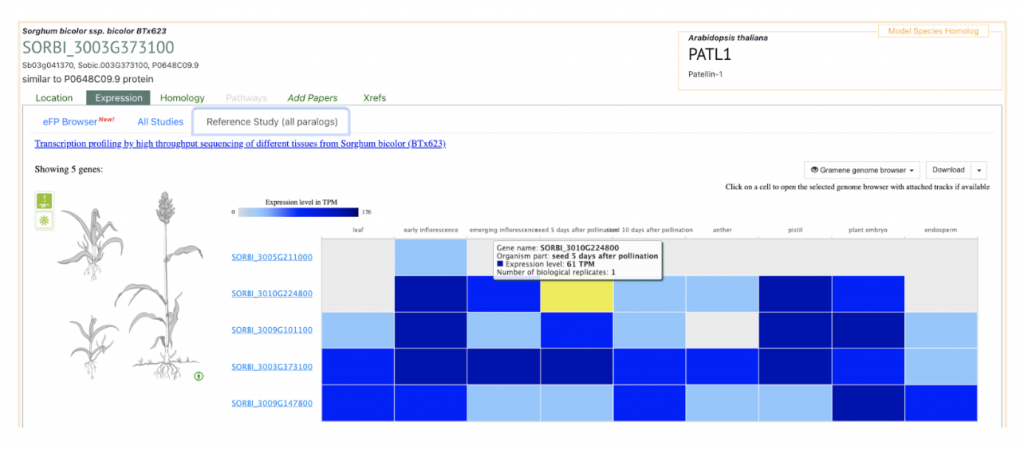
Reference:
Chaya T, Banerjee A, Rutter BD, Adekanye D, Ross J, Hu G, Innes RW, Caplan JL. The extracellular vesicle proteomes of Sorghum bicolor and Arabidopsis thaliana are partially conserved. Plant Physiol. 2023 Dec 4:kiad644. PMID: 38048422. doi: 10.1093/plphys/kiad644. Read more
Related Project Websites:
- Jeffrey Caplan’s page at the University of Delaware: https://www.udel.edu/academics/colleges/canr/departments/plant-and-soil-sciences/faculty-staff/jeffrey-caplan/
- Roger Innes’ page at Indiana University: https://biology.indiana.edu/about/faculty/innes-roger.html


