Identifying Genetic Loci for Anthracnose Resistance in Ethiopian Sorghum Germplasms: A Multi-Environment GWAS Study
Birhanu et al. identified key genetic loci associated with anthracnose resistance in Ethiopian sorghum germplasms, highlighting its potential as a valuable resource for breeding resistant sorghum varieties.
Keywords: Colletotrichum sublineola, Sorghum bicolor, Anthracnose, Disease-resistance, Genome-wide association study
This research focuses on Ethiopian sorghum landraces through multi-environment, multi-year field phenotyping to explore the genetic basis of anthracnose resistance using genome-wide association mapping. Scientists from various institutions, with diverse expertise, collaborated in identifying genetic loci associated with anthracnose resistance. The study underscores the immense value of Ethiopia’s sorghum genetic diversity in combating anthracnose, identifying key resistance loci that pave the way for advanced breeding strategies and supporting the development of resilient sorghum varieties – Birhanu
Sorghum (Sorghum bicolor), a key cereal crop in arid and semi-arid regions, is highly affected by anthracnose, a disease caused by Colletotrichum sublineola, leading to significant yield losses. Ethiopia, the center of origin and diversity for sorghum, possesses a vast germplasm collection with extensive genetic variation. Scientists from Oromia Agricultural Research Institute, Purdue University, Haramaya University, Ethiopian Institute of Agricultural Research and Agricultural Transformation Institute focused on characterizing a core subset of 358 Ethiopian sorghum accessions for anthracnose resistance using a multi-environment genome-wide association study (GWAS). Disease severity was assessed at two key growth stages, revealing that 45-56% of the accessions exhibited resistance across multiple locations. The study identified new loci associated with anthracnose resistance, highlighting the genetic diversity within Ethiopian sorghum as a valuable resource for breeding programs.
The GWAS analysis revealed significant single nucleotide polymorphisms (SNPs) associated with anthracnose resistance, with key loci identified on chromosomes 2, 4, 5 and 8. These loci were linked to genes encoding proteins such as receptor-like kinases, F-box domain proteins, and leucine-rich repeat proteins, all of which play roles in plant disease resistance. The study emphasizes the potential of Ethiopian sorghum germplasm as a source of resistance genes, which can be utilized in marker-assisted selection and breeding programs. The findings provide a foundation for further research on the genetic mechanisms of anthracnose resistance, with the goal of developing resistant sorghum cultivars that can withstand the disease and improve crop productivity in vulnerable regions.
SorghumBase examples:
SNPs strongly associated with anthracnose resistance:
- S04_66140995 -putative Receptor-Like Kinase Xa21-binding protein
- S02_75784037 – putative stress-induced antifungal kinase
- S02_2031925 – F-Box protein
Highly significant associations between anthracnose resistance and SNPs linked to genes:
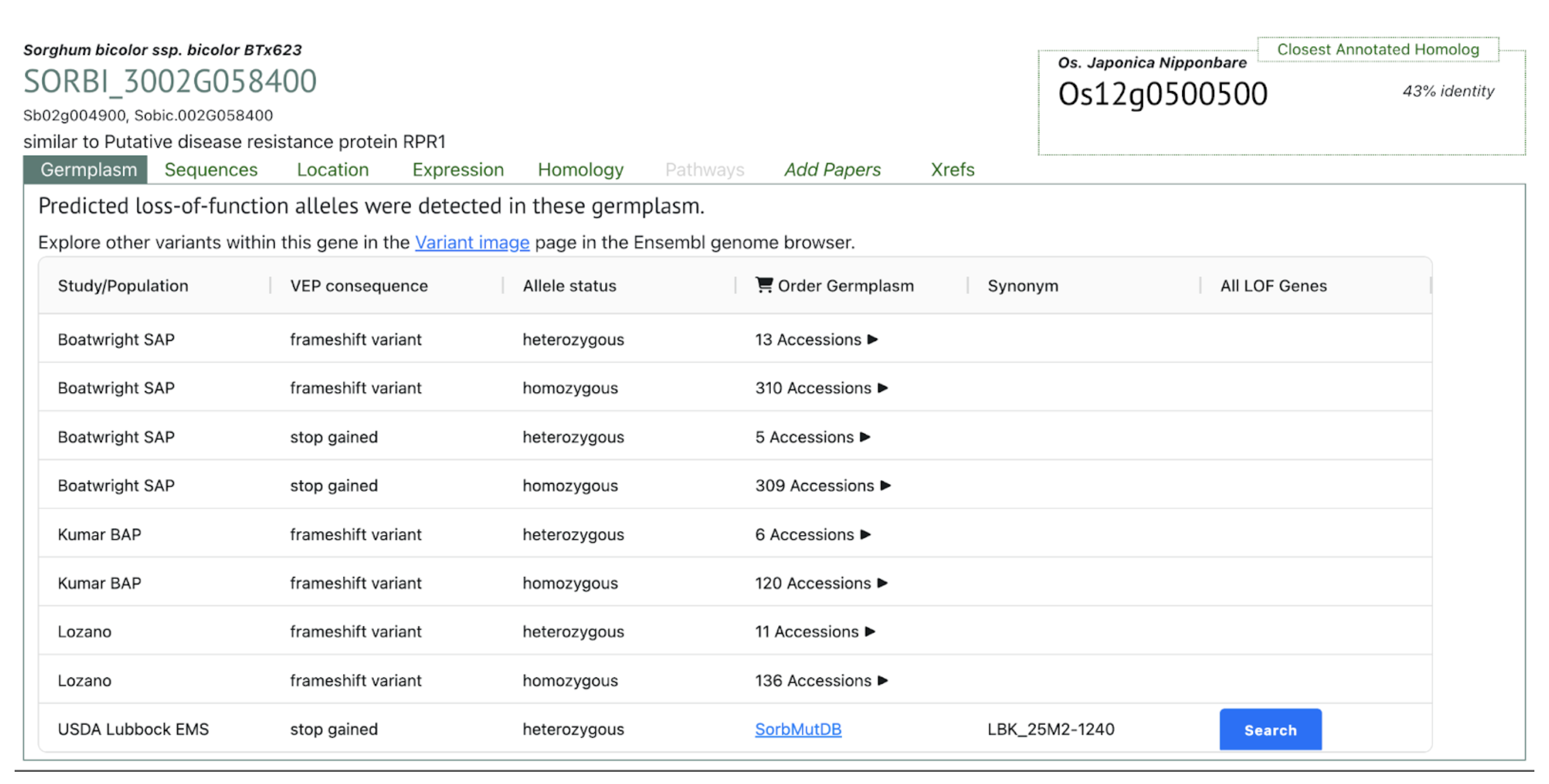
- Sobic.002G058400 – RPR1 (NLR protein) – (S02_5644024 at Sobic.002G058300)
- Sobic.008G156600 – Leucine Rich Repeat family protein
- Sobic.005G033400 – Heavy Metal Associated domain-containing protein
- Sobic.008G157400 – NB-ARC disease resistance gene

Reference:
Birhanu C, Girma G, Mekbib F, Nida H, Tirfessa A, Lule D, Bekeko Z, Ayana G, Bejiga T, Bedada G, Tola M, Legesse T, Alemu H, Admasu S, Bekele A, Mengiste T. Exploring the genetic basis of anthracnose resistance in Ethiopian sorghum through a genome-wide association study. BMC Genomics. 2024 Jul 8;25(1):677. PMID: 38977981. doi: 10.1186/s12864-024-10545-2. Read more
Related Project Websites:
- Mengiste Lab at Purdue University: https://ag.purdue.edu/department/btny/labs/mengiste/index.html



