Genome-Wide Association Mapping in Diverse Sorghum Germplasms Unveils Novel Genetic Loci for Agronomic, Root, and Stomatal Traits
Sorghum, recognized as the world’s fifth most important cereal crop, plays a pivotal role in food security owing to its resilience to various stresses and adaptability to extreme weather conditions. The study emphasized the importance of exploring the genetic basis of complex traits such as root architecture and stomatal traits, essential for enhancing abiotic stress tolerance, particularly drought stress. While most crop improvement programs focused on easily phenotyped traits, this study addressed the limitations in breeding for abiotic stress tolerance by investigating challenging traits like root morphology and stomatal traits. The authors highlighted the need for using diverse germplasm collections, such as the subset created in this study, to uncover genetic loci and candidate genes associated with these traits.
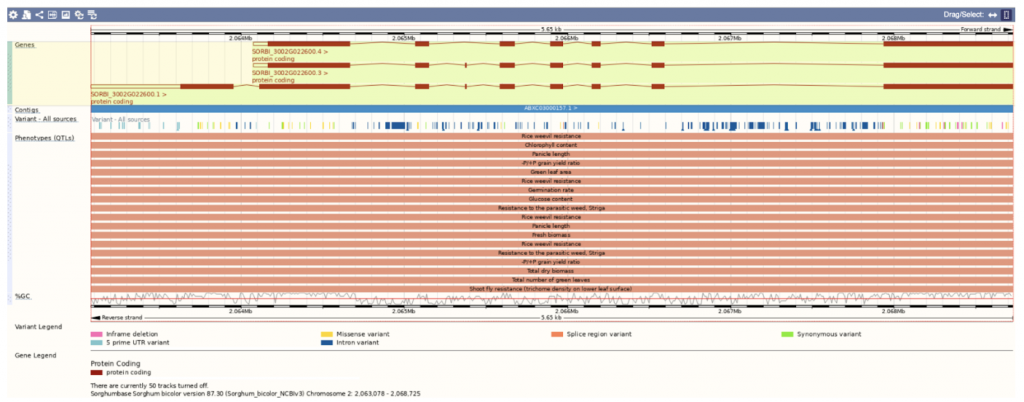
This scientific paper presents a pilot-scale genome-wide association study (GWAS) in sorghum aimed at identifying genetic loci associated with major agronomic, root morphological, and physiological traits. The study employed a subset of 96 diverse sorghum accessions, derived from a larger collection of 219 accessions, and identified 43,452 high-quality single nucleotide polymorphic (SNP) markers. The GWAS detected 42 significant quantitative trait nucleotides (QTNs) associated with various traits, including plant height, dry biomass, and stomatal density. Three novel candidate genes linked to agronomic traits were identified, including Sobic.001G499000 for plant height, Sobic.010G186600 for dry biomass, and Sobic.002G022600 for plant yield. Haplotype validation of candidate genes confirmed significant phenotypic differences, supporting their association with specific traits. The study concluded that the constructed subset of diverse sorghum lines could serve as a valuable panel for mapping key traits and accelerating molecular breeding in sorghum.
SorghumBase examples:
Reference:
Ramalingam AP, Mohanavel W, Kambale R, Rajagopalan VR, Marla SR, Prasad PVV, Muthurajan R, Perumal R. Pilot-scale genome-wide association mapping in diverse sorghum germplasms identified novel genetic loci linked to major agronomic, root and stomatal traits. Sci Rep. 2023 Dec 8;13(1):21917. PMID: 38081914. doi: 10.1038/s41598-023-48758-2. Read more
Related Project Websites:
- Ramasamy Perumal’s Lab/ Sorghum Breeding at Kansas State University: https://www.hays.k-state.edu/programs/sorghum/index.html
- Raveendran Muthurajan’s page at Tamil Nadu University: https://tnau.ac.in/site/research/directorss-desk/

