Functional Diversity of CYP79A Genes in Sorghum: Roles in Metabolism, Defense, and Adaptation
This study explores the functional diversity of sorghum CYP79A genes, revealing their roles in amino acid metabolism, plant defense, growth regulation, and environmental adaptation.
Keywords: CYP79A, oximes, tissue-specific expression, plant signaling
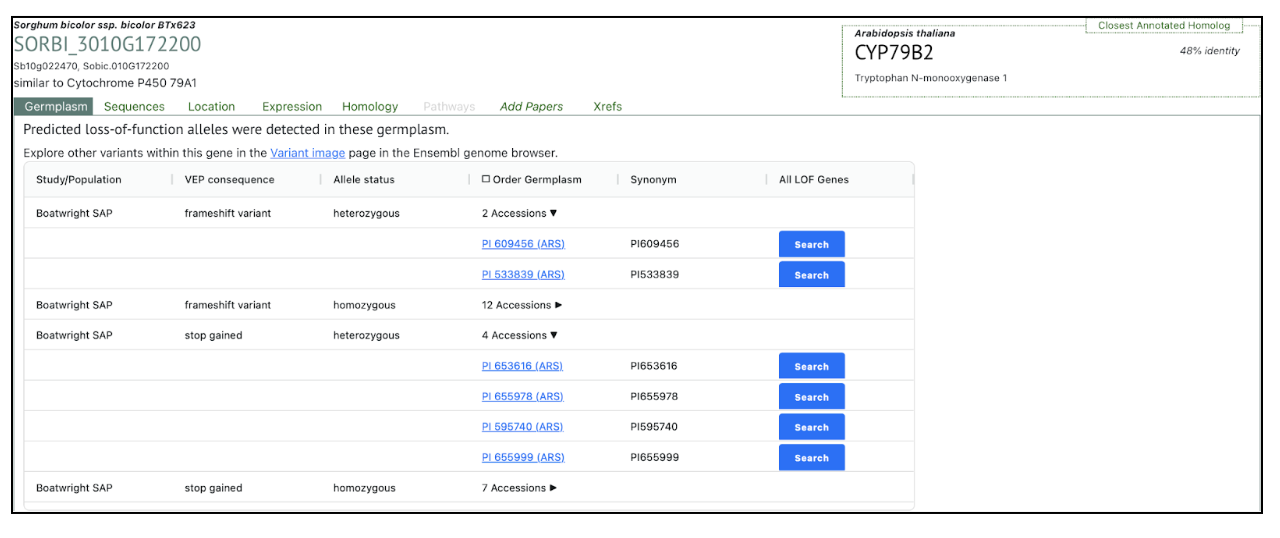
In this collaborative study with Professor Mullet’s group we are happy to bring to life CYP79A genes in sorghum previously considered to be silent. In future studies we aim to study the functions of the individual CYP79A genes in sorghum taking advantage of the FIND-IT technology in producing knock-out variants of the individual CYP79A- genes. The SorghumBase is really helpful to us, e.g. by rapidly guiding us to new CYP79A mutants and updating us on exciting new discoveries from around the entire sorghum world. – Møller
In a publication in 2018 in Molecular Plant, we designated oximes and unrecognized chameleons in general and specialized plant metabolism. The current study represents a step forward in the elucidation of their multiple functions in plants. I am really happy to be part of this highly interdisciplinary study and happy that we were able to use a future crop plant as the experimental system instead of a model plant. – Sørensen
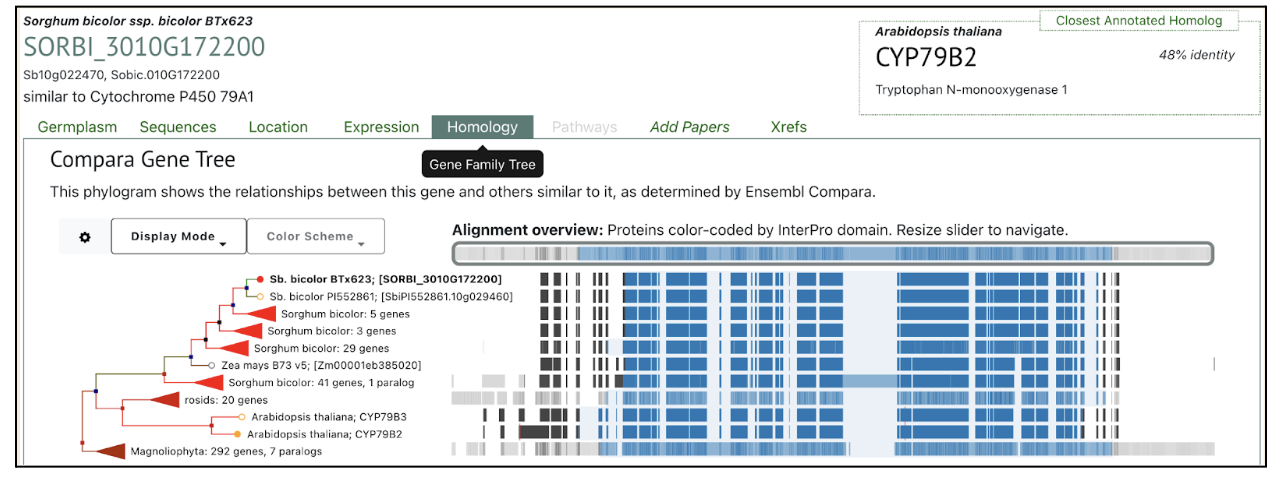
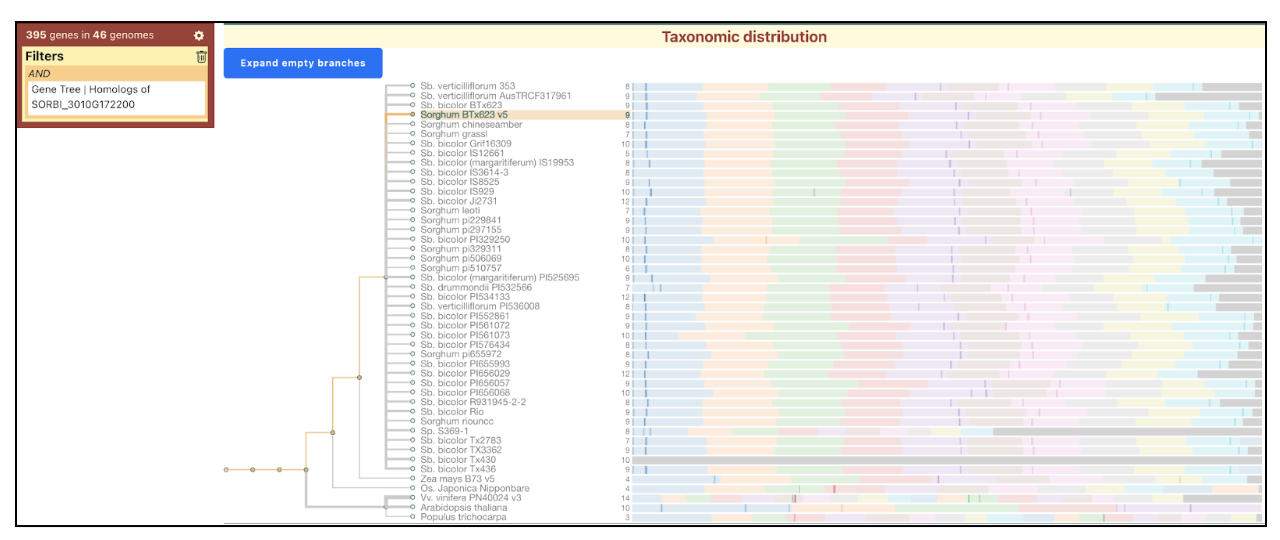
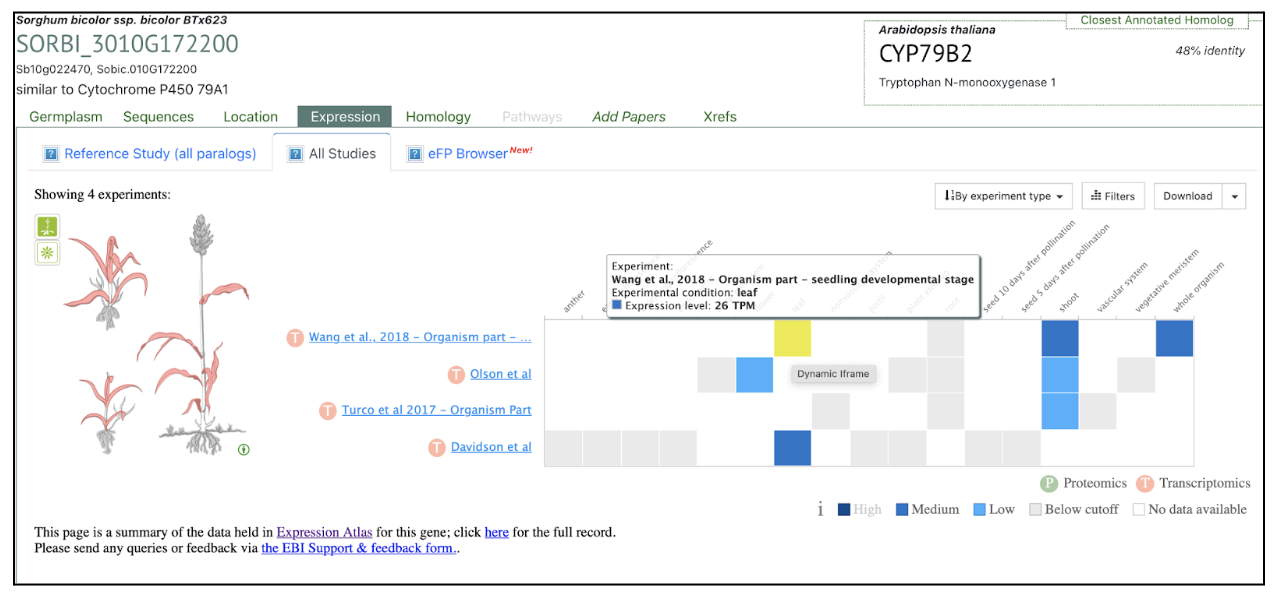
Scientists from University of Copenhagen and Texas A&M University investigated the functional diversity of CYP79A genes in sorghum, focusing on their role in amino acid metabolism and cyanogenic glucoside biosynthesis. While CYP79A1 is well-established as a key enzyme in dhurrin production, this research characterizes the functions of eight additional CYP79A genes. Phylogenetic analysis segregates these genes into two distinct clades. SbCYP79A61, a member of Clade A, shares enzymatic similarities with maize orthologues, catalyzing the conversion of phenylalanine into phenylacetaldoxime. Functional assays reveal that Clade B members exhibit varied substrate specificities, with CYP79A91, CYP79A93 and CYP79A95 converting valine and isoleucine to their respective oximes, whereas CYP79A94 only converts isoleucine. CYP79As are involved in producing volatile oximes that may function in plant signaling and herbivore interactions. Further, CYP79A93 also converts phenylacetaldoxime, however, seemingly to a lesser extent than SbCYP79A61. The expression of these genes is highly tissue-specific, with CYP79A61 predominantly expressed in mature leaves, hinting at a role in plant defense mechanisms.
The study also suggests that CYP79A-derived oximes and their metabolic derivatives contribute to plant growth regulation, defense, and environmental adaptability. Expression patterns suggest potential involvement in root architecture, flowering regulation, and stress responses, such as drought tolerance. The presence of CYP79A genes in other monocots implies a broader evolutionary significance. Future studies using gene knock-out techniques and yeast-based assays aim to further clarify their physiological roles. Overall, this research highlights the functional complexity of CYP79A enzymes beyond dhurrin biosynthesis, emphasizing their importance in sorghum’s adaptive strategies and metabolic networks.
SorghumBase examples:
Koleva DT, Liu M, Dusak B, Ghosh S, Krogh CT, Hellebek IR, Cortsen MT, Motawie MS, Jørgensen FS, McKinley BA, Mullet JE, Sørensen M, Møller BL. Amino acid substrate specificities and tissue expression profiles of the nine CYP79A encoding genes in Sorghum bicolor. Physiol Plant. 2025 Jan-Feb;177(1):e70029. PMID: 39749417. doi: 10.1111/ppl.70029. Read more
Related Project Websites:
- Dr. Møller’s page at the University of Copenhagen: https://synbio.ku.dk/about/blm/
- Dr. Mullet’s page at Texas A&M University: https://bcbp.tamu.edu/people/mullet-john/





