Ex-PVP Sorghum Genomes: A Treasure Trove for Breeding and Functional Genomics
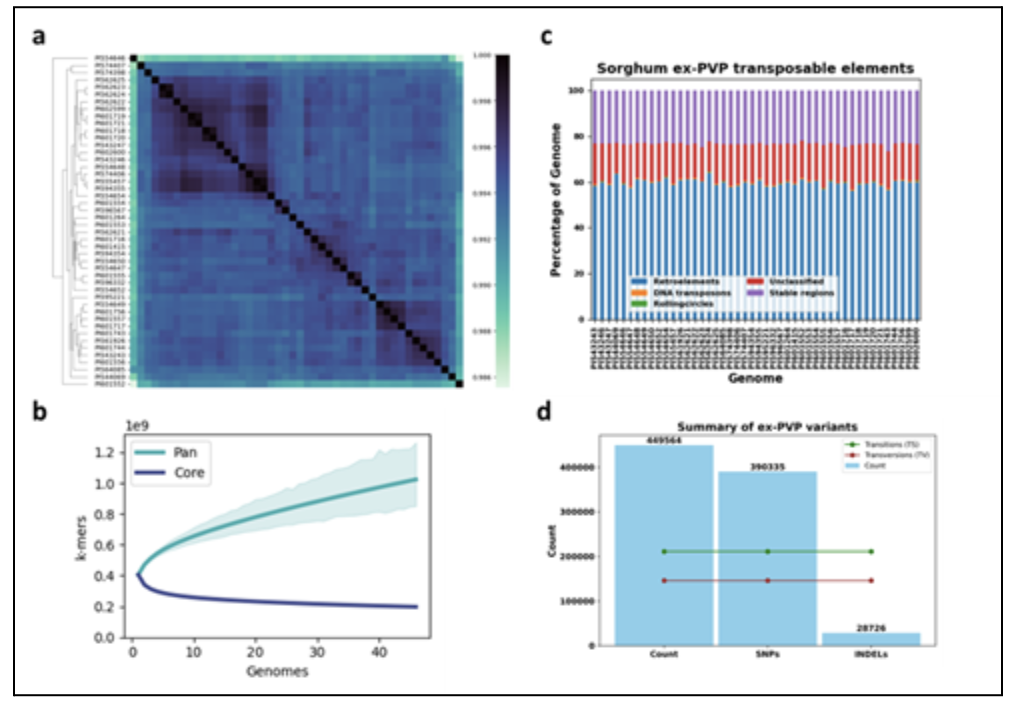
Sorghum bicolor, a diploid cereal crop with a genome size of approximately 730 Mb and 10 haploid chromosomes, is the fifth most produced cereal worldwide (Paterson et al., 2009). Primarily cultivated in Africa, sorghum is also recognized for its potential as a biofuel crop and a source of cellulosic feedstock (George et al., 2022). Its relatively compact genome makes sorghum an excellent model for functional genomics studies of Saccharinae and other C4 grasses. In the United States, Plant Variety Protection (PVP) laws safeguard inbred sorghum lines developed by private companies for 20 years. After this period, these lines, known as ex-PVPs, become publicly accessible, providing valuable genetic resources for breeding programs aimed at enhancing yield, disease resistance, and climate resilience. A team led by Dr. Michael Todd at the Salk Institute assembled and analyzed 46 new Sorghum bicolor ssp. bicolor ex-PVP breeding lines. Seeds were obtained from the USDA Germplasm Information Resource Network (GRIN). Pairwise genomic comparisons of these genomes show high nucleotide identity with minor clustering with transposable elements constituting around 60% of the genome (Figure 1).
In addition, Cold Spring Harbor Laboratory (CSHL) mapped 66,928 Sorghum pan-genes developed at CSHL using annotations from 28 sorghum genomes were lifted to each of the above ex-PVP accessions using the Liftoff tool. The lifted pan-gene annotations are provided as a track on the corresponding ex-PVP browser pages. Approximately 41 million Reference SNP cluster identifiers (rsIDs) assigned by the European Variation Archive (EVA) were mapped to these genomes using EVA variant mapping pipeline.

References:
- Paterson, Andrew H., John E. Bowers, Rémy Bruggmann, Inna Dubchak, Jane Grimwood, Heidrun Gundlach, Georg Haberer, et al. 2009. “The Sorghum Bicolor Genome and the Diversification of Grasses.” *Nature* 457 (7229): 551–56.
- George, Toyosi T., Anthony O. Obilana, Ayodeji B. Oyenihi, Anthony B. Obilana, Damilola O. Akamo, and Joseph M. Awika. 2022. “Trends and Progress in Sorghum Research over Two Decades, and Implications for Global Food Security.” *South African Journal of Botany: Official Journal of the South African Association of Botanists = Suid-Afrikaanse Tydskrif Vir Plantkunde: Amptelike Tydskrif van Die Suid-Afrikaanse Genootskap van Plantkundiges* 151 (December): 960–69.

