Comprehensive Transcriptomic Atlas and Co-Expression Networks Reveal Gene Regulatory Mechanisms in Sorghum
In order to build a resource for the comprehensive study of gene expression in sorghum, researchers from Saint Louis University, the USDA-ARS and the Chinese Academy of Sciences analyzed 873 RNA-sequencing datasets from 19 tissues, resulting in the creation of the most detailed transcriptomic atlas for this crop to date. This atlas revealed key insights into the gene expression landscape, including the identification of 595 housekeeping genes (HKGs) and 2080 tissue-specific expression genes (TEGs). HKGs, which are ubiquitously expressed across all tissues, exhibited higher expression levels and stronger selective constraints compared to TEGs, which are highly specialized and evolve more rapidly. The constructed transcriptome-wide co-expression network (TW-CEN) comprised 35 modules enriched with specific Gene Ontology terms, highlighting the relationships between gene connectivity, expression levels, and evolutionary pressures. This extensive transcriptomic resource will aid functional characterization and genomic studies in sorghum, particularly in understanding complex agricultural traits and regulatory mechanisms.
Additionally, the study delved into the starch synthesis pathways, revealing the integral roles of photosynthesis and microtubule-based movement. A starch synthesis pathway co-expression network (SSP-CEN) was constructed using 102 guide genes, which were categorized into four expression patterns across various tissues, especially seed-related ones. Notably, the SSP-CEN identified critical modules containing a significant portion of the starch pathway genes, underscoring their importance in starch biosynthesis. This network-based approach provides a framework for future research on the regulatory mechanisms of starch synthesis in sorghum, facilitating the identification of candidate genes for crop improvement. Overall, this study significantly enhances the functional genomics toolkit available for sorghum, paving the way for advances in crop yield and stress adaptation through informed breeding strategies.
SorghumBase examples:
The highest expressed gene Sobic.008G043200, log2(TPM+1) = 19.90) from this published study has been selected as the candidate gene to explore SorghumBase (https://sorghumbase.org/).
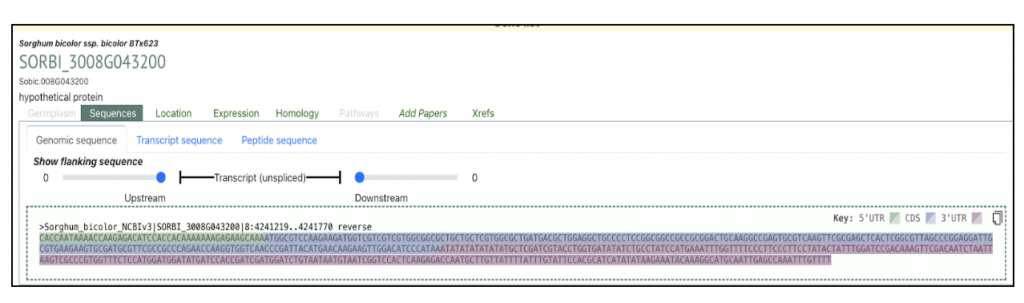
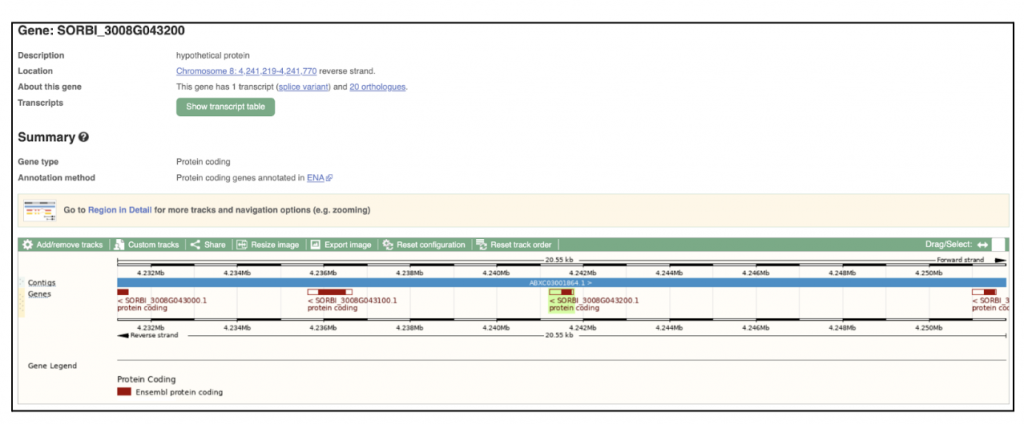
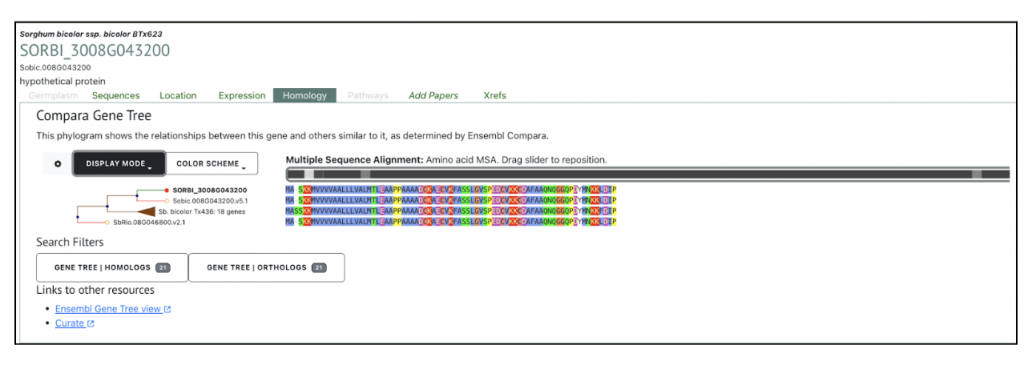
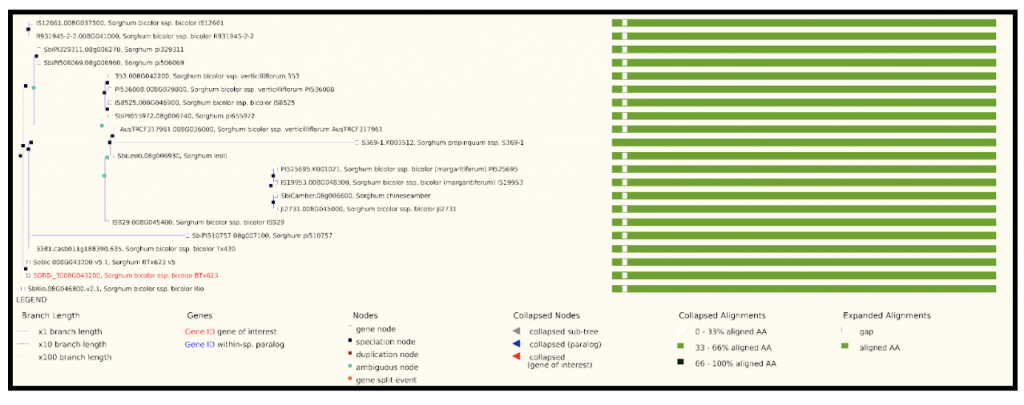
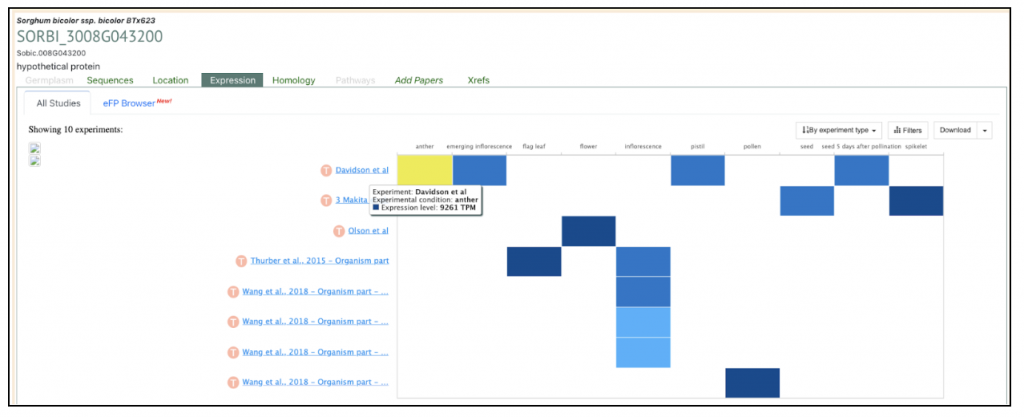
Reference:
Hu Z, Chen J, Olatoye MO, Zhang H, Lin Z. Transcriptome-wide expression landscape and starch synthesis pathway co-expression network in sorghum. Plant Genome. 2024 Apr 11:e20448. PMID: 38602082. doi: 10.1002/tpg2.20448. Read more
Related Project Websites:
- Lin lab at St. Louis University: http://www.zlinlab.org/

