Accelerating Gene Discovery in Sorghum: EMS Mutagenesis and Sequencing Reveal a Conserved Regulator of Male Fertility
Researchers demonstrated how coupling EMS mutagenesis with sequencing accelerates gene identification and validation in sorghum, revealing a conserved bHLH transcription factor essential for male fertility and highlighting broader applications for crop improvement and functional genomics.
Keywords: bulked segregant analysis, chemical mutagenesis, germinal sectors, male sterility, non-complementation, sorghum
One of the limitations in gene function discovery is the recovery of additional alleles to demonstrate the molecular identity of the gene responsible for phenotype expression. We identified sectors displaying the phenotype on mutagenized heterozygous plants and discovered the gene responsible by finding the newly induced allele in whole genome sequencing data. This approach doesn’t have to wait for the next generation to identify genes from phenotypes and can rapidly leverage single alleles in the reverse genetics resources of sorghum to forward genetic discovery engines. – Dilkes
If we are going to meet the growing demands of world food security and sustainable bioenergy production, we need more efficient ways to study the functions of genes and how they are controlled directly in crop systems. Bottlenecks in plant transformation currently limit many researchers from the benefits of gene editing tools. The targeted EMS mutagenesis approach described here highlights a more accessible alternative in recalcitrant crop systems. – Eveland
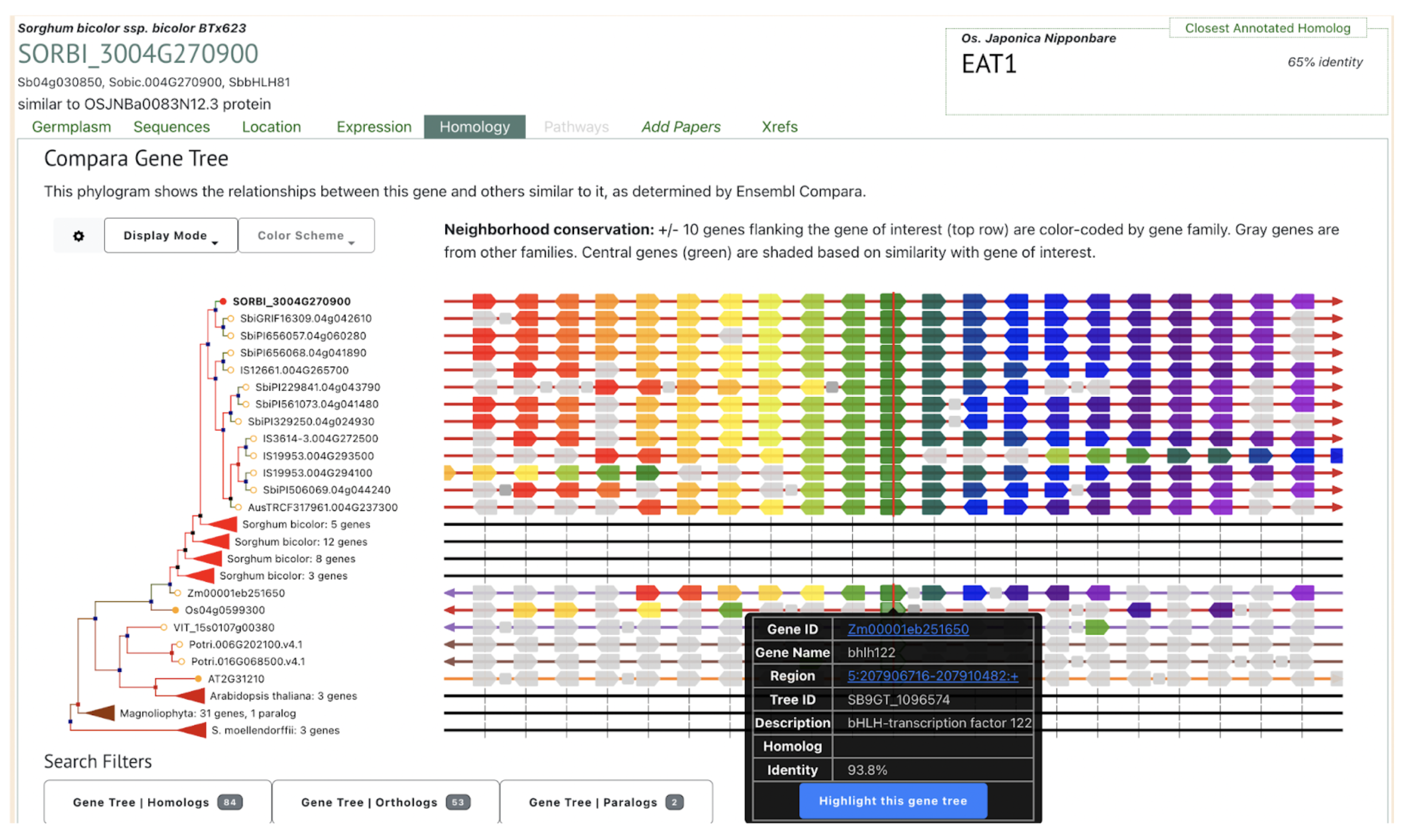
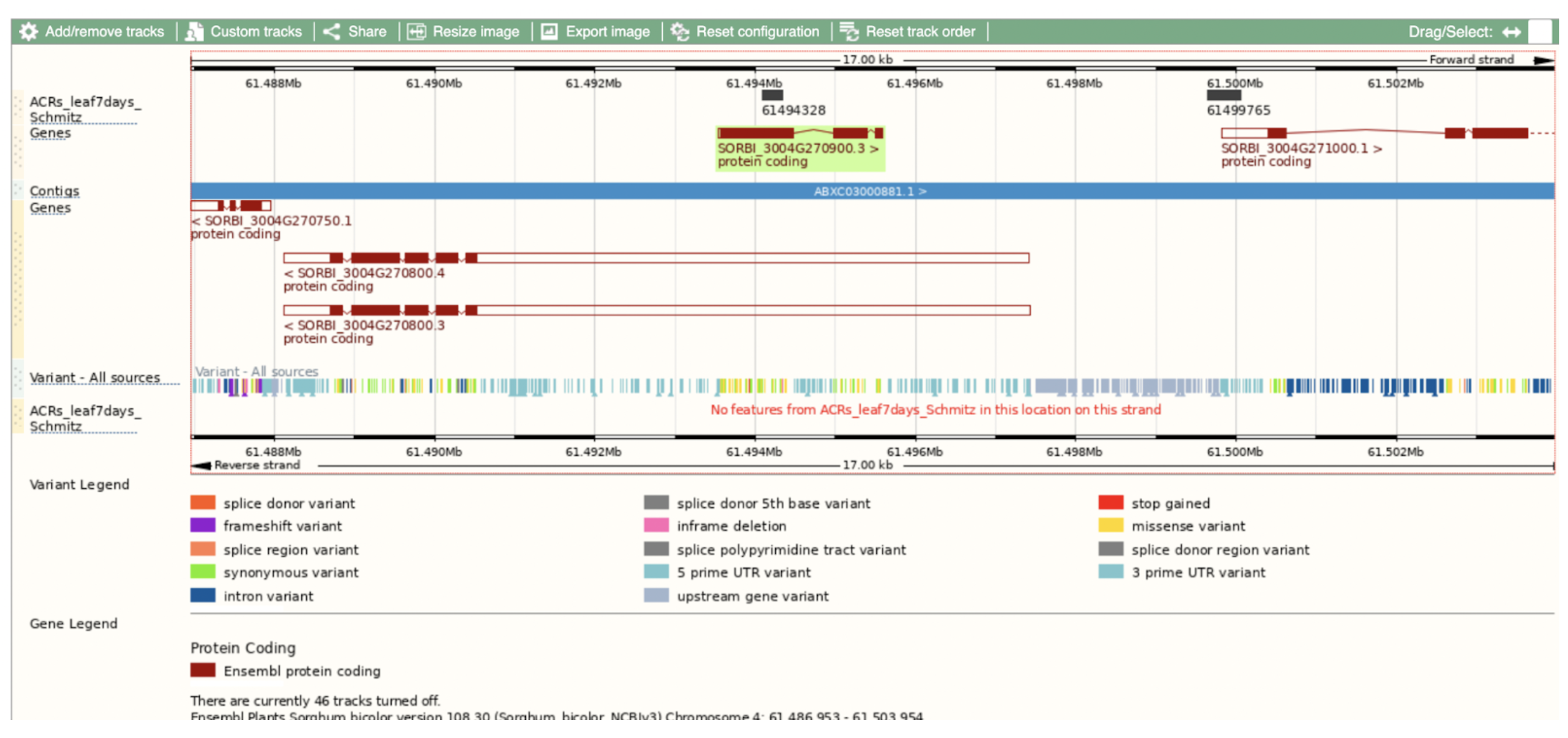
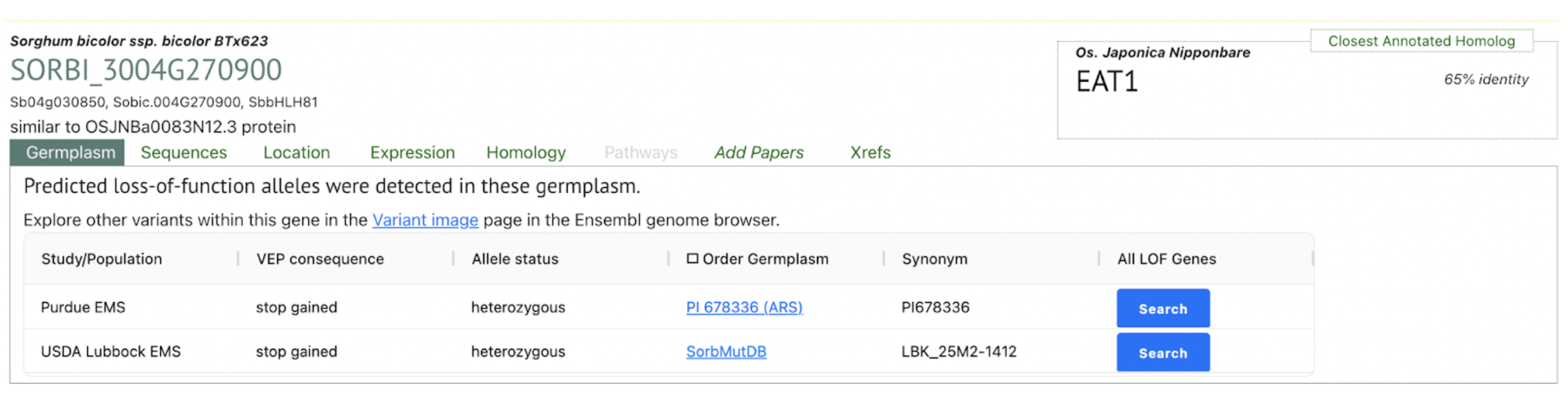
Ethyl methanesulfonate (EMS) mutagenesis is a well-established tool for generating mutant alleles in functional genomics studies. However, gene validation often requires independent alleles that fail to complement a phenotype, which can delay research progress. Scientists from Donald Danforth Plant Science Center and Purdue University demonstrated how coupling classical EMS mutagenesis with sequencing can accelerate gene identification and validation in sorghum, an important but genetically challenging crop. The research identified a conserved bHLH transcription factor required for male fertility, responsible for the sorghum ms8 phenotype. Given the regulatory and technical challenges associated with genetic transformation in sorghum, targeted mutagenesis provides an efficient alternative for developing novel alleles in desired genetic backgrounds. This approach reduces the need for introgressing mutations from unrelated backgrounds and simplifies breeding by generating mutations directly in elite lines. Additionally, it offers a practical solution for researchers who lack access to transformation facilities, enabling broader applications in plant breeding and functional genetics.
Beyond sorghum, the study highlights the conservation of bHLH transcription factors in male sterility regulation across plant species, including maize. The ms8-1 allele, caused by a nonsense mutation in the sorghum homolog of maize bhlh122, disrupts tapetum development, a critical process for pollen maturation. The functional conservation of this pathway across monocots and dicots suggests potential applications in hybrid seed production and crop improvement. Moreover, EMS mutagenesis and sequencing strategies could be extended to long-lived and clonally propagated species, where traditional breeding is impractical. By leveraging mitotically cohesive sectors, this approach enables faster gene function discovery, expanding the utility of mutagenesis for species with long generation times and limited genetic transformation resources.
SorghumBase examples:
Xiao Y, Khangura RS, Wang Z, Dilkes BP, Eveland AL. Targeted seed EMS mutagenesis reveals a basic helix-loop-helix transcription factor underlying male sterility in sorghum. Genetics. 2025 Jul 9;230(3):iyaf017. PMID: 39882980. doi: 10.1093/genetics/iyaf017. Read more
Related Project Websites:
- Eveland Lab at Donald Danforth Plant Science Center: https://www.danforthcenter.org/our-work/principal-investigators/andrea-eveland/
- Dr. Dilkes’ page at Purdue University: https://ag.purdue.edu/directory/bdilkes




