Sorghum Gene Locus Confers Resistance to Colletotrichum sublineola and Other Fungi
Sorghum is the fifth most popular cereal crop world-wide and is a key crop for many people in Asia and Africa. Globally, there is a 15% – 30% crop loss due to pathogens. Anthracnose, a disease prevalent in sorghum around the world, is caused by the fungus Colletotrichum sublineola (Cs). Selective breeding for resistance to specific disease has been successful with some crops and is environmentally friendly. Scientists, breeders and growers have great interest studying genetic resistance to limit disease and reduce crop loss.
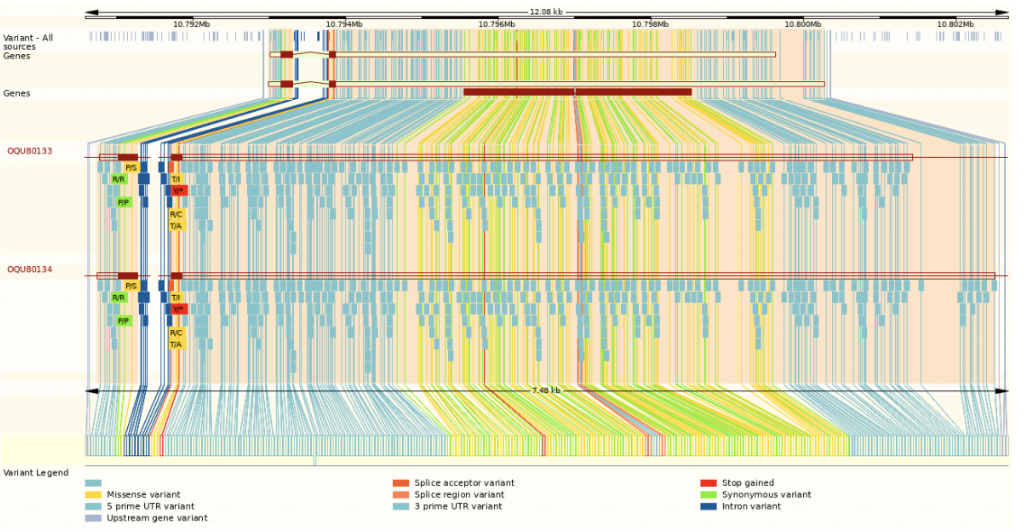
In an effort to better understand regulators of specific resistance, researchers from Purdue University screened sorghum varieties for Cs resistance. The researchers discovered that the sorghum genotype SC283 showed resistance to several strains of Cs, while the genotype TAM428 lacks such resistance. Recombinant inbred lines (RILs) were produced by crossing these two lines that were clearly either resistant or susceptible, and whole-genome resequencing showed a locus that confers Cs resistance as well as resistance to other fungi. Researchers discovered a major fungal resistance locus in sorghum composed of the nucleotide-binding leucine-rich repeat receptor (NLR) gene ANTHRACNOSE RESISTANCE GENE1 (ARG1) that is completely nested in an intron of a cis-natural antisense transcript (NAT) gene designated CARRIER OF ARG1 (CARG). DNA- and RNA-Seq analysis showed that resistant RILs had a loss of CARRIER OF ARG1 (CARG) transcription and contained miniature inverted-repeat transposable element (MITE) insertions in the ARG1 promoter region. These were associated with an increase in the expression of ARG1 which in turn was associated with higher histone H3K4 and H3K36 methylation and resistance to a wide variety of fungal pathogens. These pathogens include Puccinia purpurea (an obligate biotrophic fungus), Cs (a hemibiotrophic fungus) and Bipolaris sorghicola (a necrotrophic fungus); such broad resistance is rare. Conversely, the non-resistant RILs express CARG and produce two ARG1 transcripts which encode truncated NLR proteins resulting accompanied by enhanced expression of the natural antisense transcripts (NATs).This reduced expression of ARG1 is associated with lower histone H3K4 and H3K36 methylation and susceptibility to infection. The CARG–ARG1 locus shows great promise for sorghum improvement as it can be introgressed into cultivars to develop lines that are widely resistant to fungal pathogens.
In addition to the exciting studies that can be pursued to untangle the biological and molecular mechanisms of how CARG-ARG1 locus function, this work has significant practical implications for breeding disease resistant sorghums though traditional breeding or biotechnological applications. – Mengiste and Lee
SorghumBase Example:
Reference:
Lee S, Fu F, Liao CJ, Mewa DB, Adeyanju A, Ejeta G, Lisch D, Mengiste T. Broad-spectrum fungal resistance in sorghum is conferred through the complex regulation of an immune receptor gene embedded in a natural antisense transcript. Plant Cell. 2022 Apr 26;34(5):1641-1665. PMID: 35018449. DOI: 10.1093/plcell/koab305. Read more
Related Project Websites:
Purdue University page: https://www.purdue.edu/gradschool/pulse/groups/profiles/faculty/mengiste.html
Figure 1. Sanghun Lee, research associate in the Mengiste Lab and first co-author of the study, holding a petri plate with Colletotrichum sublineola culture. Photo credit Tom Campbell, Purdue University.

Figure 2. Sorghum genotypes that are resistant (left) or susceptible (right) to C. sublineola in Western Ethiopia, Jimma, under natural pathogen infection in the field. Photo credit Tesfaye Mengiste.

