Role of CcNAC6 in Regulating Secondary Cell Wall Synthesis and Lignin Content in Sudan Grass
Sudan grass (Sorghum sudanense S.), a high-quality sorghum forage,predominantly stores its lignocellulose in the secondary cell wall. The structural carbohydrates in the cell wall of Sudan grass, including cellulose, hemicellulose, and lignin, are essential nutrients for ruminants like cattle and sheep. Microorganisms in the rumen digest cellulose and hemicellulose, while lignin remains indigestible and hinders the digestion of other nutrients. Therefore, reducing lignin content to improve the digestibility of forage is crucial. This goal is e achieved by understanding the genetic basis of secondary cell wall synthesis. Transcriptome sequencing has identified CcNAC1 as a regulator of secondary wall synthesis in Sudan grass, but comprehensive analysis of other CcNAC genes in Sudan grass is lacking . Researchers from the Jiangsu Academy of Agricultural Sciences and Nanjing Agricultural University performed a bioinformatics analysis which revealed that CcNAC6 is highly homologous to AtSND2 in Arabidopsis and can increase lignin content when overexpressed, indicating its potential role in enhancing lignin synthesis and impacting forage quality.
Further bioinformatics and phylogenetic analysis showed that among the nine CcNAC genes, only CcNAC6 and CcNAC8 have high homology with AtSND2 and AtVND2, respectively. These genes are associated with secondary cell wall synthesis. The analysis also indicated that NAC transcription factors, like CcNAC6, may regulate secondary wall synthesis directly or indirectly through interactions with other proteins, such as cysteine proteases involved in programmed cell death (PCD). Additionally, promoter element analysis suggests that CcNAC gene expression in Sudan grass is influenced by light and hormones such as abscisic acid, jasmonic acid, and brassinosteroids. The study demonstrated that CcNAC6 could be a key regulator of secondary cell wall synthesis, with potential applications in breeding strategies aimed at developing Sudan grass varieties with lower lignin content for better digestibility and economic benefits in agriculture.
SorghumBase example:
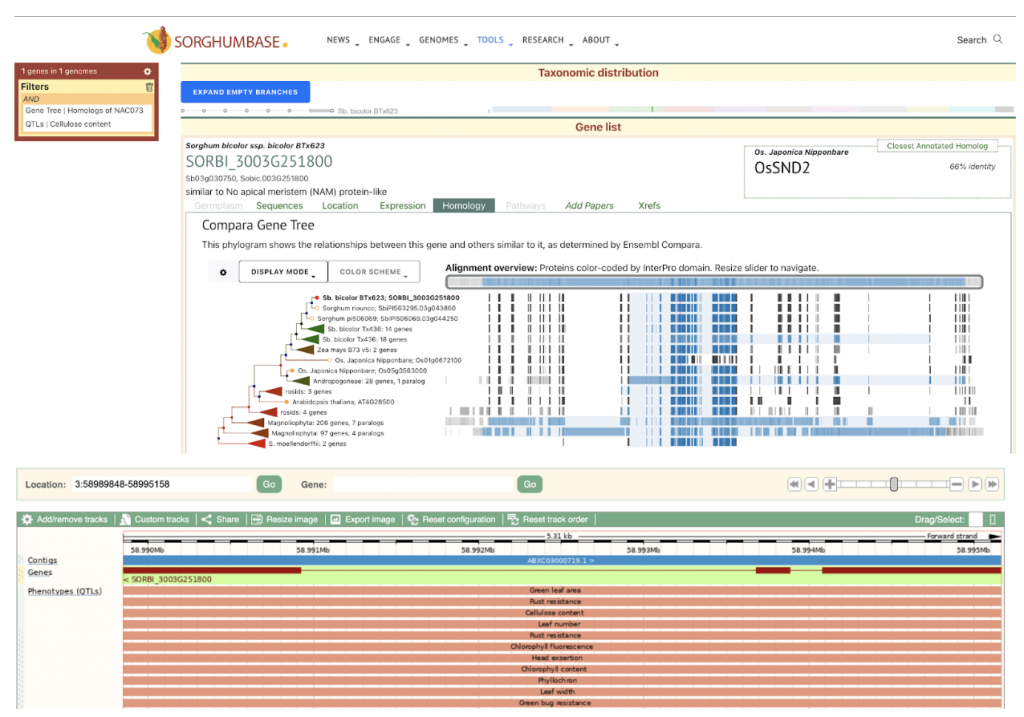
Sorghumbase does not host the sudan grass genome, but it is possible to explore the gene family using Sorghumbase. Sorghum bicolor orthologs of AtSND2 are SORBI_3009G231600 and SORBI_3003G251800. Overlapping QTLs can be used to limit gene search results and can be seen in the ensembl genome browser. The Sorghum QTL Atlas includes a cellulose content QTL that overlaps the SORBI_3003G251800 locus.
Reference:
Huang Y, Wu J, Lin J, Liu Z, Mao Z, Qian C, Zhong X. CcNAC6 Acts as a Positive Regulator of Secondary Cell Wall Synthesis in Sudan Grass (Sorghum sudanense S.). Plants (Basel). 2024 May 14;13(10):1352. PMID: 38794423. doi: 10.3390/plants13101352. Read more

