QTLs Identified for Sorghum Resistance to Illinois C. sublineola Pathotype
Anthracnose leaf blight (ALB) is a fungal disease caused by Colletotrichum sublineola that results in significant economic loss for sorghum crops. Mitigating the effects of this pathogen is achieved mostly through plant resistance. However, this resistance can differ depending on the pathogen pathotype (a classification of different strains of a pathogen based on their virulence to plants with specific resistance genes). Discovering resistance to a broader range of C. sublineola pathotypes is of great interest to breeders, growers, and scientists.
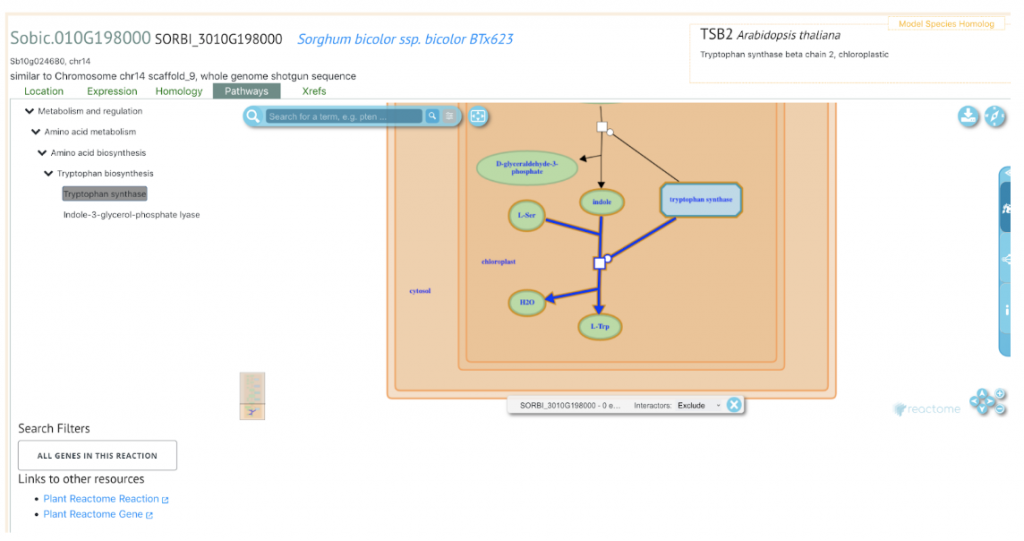
In an effort to locate loci associated with resistance to a unique pathotype of C. sublineola, Khanal and colleagues at the University of Illinois at Urbana-Champaign isolated the fungus from infected leaves in Illinois. This unique Illinois pathotype (19CS002.1) was used to inoculate 579 temperate-adapted sorghum conversion lines in 2019 and 2020 so that the resistant plants could be further evaluated using the genome-wide association study (GWAS) and a metabolic pathway analysis using the Pathway Associated Study Tool (PAST). The researchers identified 47 markers associated with resistance to C. sublineola across all of the chromosomes except chromosome 8. Of these, 32 genes were selected as likely candidates due to their location near significant SNPs. For example, the significant SNPs on chromosomes 5 and 10 were in close physical proximity to those determined in previous studies to be associated with anthracnose resistance to isolates from Puerto Rico and Texas (Cuevas et al. 2018; Prom et al. 2019). These chromosomal regions likely impart a broader resistance to anthracnose across a variety of environments. Candidates genes from these chromosomes include Sobic.005G063200 (chrom. 5) and Sobic.010G198000 (chrom. 10) both of which have orthologs associated with Tryptophan pathways in rice and Arabidopsis respectively. This coincides with research which found that sorghum lines with high levels of tryptophan showed greater resistance to C. sublineola (Tugizimana et al., 2019). The scientists also pinpointed candidate genes on chromosomes 2 and 3 (Sobic.002G247500, Sobic.003G393400) the orthologs of which have been shown to play a role in bacterial and fungal resistance in Arabidopsis (Chen et al., 2009; Pečenková et al., 2011; Stegmann et al., 2012).
In addition, based on the combined results of the GWAS and PAST, 47 metabolic pathways were associated with C. sublineola resistance. The analysis also indicated that secondary metabolites likely also have a part to play in resistance. Furthermore, a shikimate O-hydroxycinnamoyltransferase gene involved in the phenylpropanoid pathway was associated with sorghum anthracnose resistance in this study indicating that there is functional genetic variation in various biosynthetic pathways.
“Plant pathogen variants pose challenges for disease management through host resistance. We evaluated a sorghum genetic resource against a local strain of the pathogen and found novel as well as conserved associations with resistance. Our results should help to develop varieties resistant to local pathogens in Illinois and to identify conserved mechanisms of defense that are useful anywhere sorghum is grown.” – Mideros
SorghumBase example:
References
Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, Guo H, Zhou JM. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009 Aug;21(8):2527-40. PMID: 19717619. DOI:: 10.1105/tpc.108.065193. Read more
Cuevas HE, Prom LK, Cooper EA, Knoll JE, Ni X. Genome-Wide Association Mapping of Anthracnose (Colletotrichum sublineolum) Resistance in the U.S. Sorghum Association Panel. Plant Genome. 2018 Jul;11(2). PMID: 30025025. DOI: 10.3835/plantgenome2017.11.0099. Read more
Khanal A, Adhikari P, Kaiser C, Lipka AE, Jamann TM, Mideros SX. Genetic mapping of sorghum resistance to an Illinois isolate of Colletotrichum sublineola. Plant Genome. 2022 Jul 13:e20243. PMID: 35822435. DOI: 10.1002/tpg2.20243. Read more
Pecenková T, Hála M, Kulich I, Kocourková D, Drdová E, Fendrych M, Toupalová H, Zársky V. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J Exp Bot. 2011 Mar;62(6):2107-16. PMID: 21199889. DOI:: 10.1093/jxb/erq402. Read more
Prom LK, Ahn E, Isakeit T, Magill C. GWAS analysis of sorghum association panel lines identifies SNPs associated with disease response to Texas isolates of Colletotrichum sublineola. Theor Appl Genet. 2019 May;132(5):1389-1396. PMID: 30688991. DOI: 10.1007/s00122-019-03285-5. Read more
Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Žársky V, McDowell JM, Shirasu K, Trujillo M. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell. 2012 Nov;24(11):4703-16. PMID: 23170036. DOI: 10.1105/tpc.112.104463. Read more
Tugizimana F, Djami-Tchatchou AT, Steenkamp PA, Piater LA, Dubery IA. Metabolomic Analysis of Defense-Related Reprogramming in Sorghum bicolor in Response to Colletotrichum sublineolum Infection Reveals a Functional Metabolic Web of Phenylpropanoid and Flavonoid Pathways. Front Plant Sci. 2019 Jan 4;9:1840. PMID: 30662445. DOI: 10.3389/fpls.2018.01840. Read more
Related Project Websites:
Jamann Lab: https://publish.illinois.edu/tjamann/
Mideros Lab: http://publish.illinois.edu/fieldcroppathologylab/





