Mechanisms of Aphid Resistance in Sorghum: The Role of Leaf Structure and Flavonoid Biosynthesis
Sorghum (Sorghum bicolor L.) is a critical crop grown globally, with high adaptability to arid environment. However, its yield and quality are threatened by the sorghum aphid (Melanaphis sacchari), which infests the plant during its late growth phase. Scientists from Hebei Agricultural University, and North China Key Laboratory for Crop Germplasm Resources of Education Ministry conducted a study which focused on investigating the mechanisms behind sorghum’s resistance to aphid infestation by comparing aphid-resistant (HN16) and aphid-susceptible (BTx623) sorghum varieties. The research involved examining the leaf epidermal cell structure and conducting transcriptome and metabolome analyses. Findings indicated that aphid resistance in sorghum correlates with more regular epidermal cells and reduced intercellular spaces and leaf thickness. Transcriptome and metabolome analyses revealed that the differentially expressed genes (DEGs) and metabolites in these varieties are primarily involved in the flavonoid biosynthesis pathway, with naringenin and genistein significantly upregulated in response to aphid infestation, suggesting their vital roles in plant defense.
Further experiments confirmed that exogenous applications of naringenin and genistein at 0.1% concentration enhance aphid resistance in sorghum. These findings underscore the importance of flavonoid biosynthesis in the plant’s defensive response and highlight the potential for developing aphid-resistant sorghum varieties through genetic and metabolic pathway manipulations.
SorghumBase examples:
The gene IDs mentioned in the study include https://sorghumbase.org/genes?idList=SORBI_3002G117900,SORBI_3004G166700,SORBI_3006G160700,SORBI_3010G072300,SORBI_3010G276700,SORBI_3004G357600,SORBI_3001G378300,SORBI_3001G344500,SORBI_3001G146400,SORBI_3001G146500,SORBI_3004G236300,SORBI_3001G427700. These gene IDs are associated with various metabolic pathways, including starch and sucrose metabolism, IAA biosynthesis, IAA signal transduction pathway, and Trp biosynthesis pathway, among others.
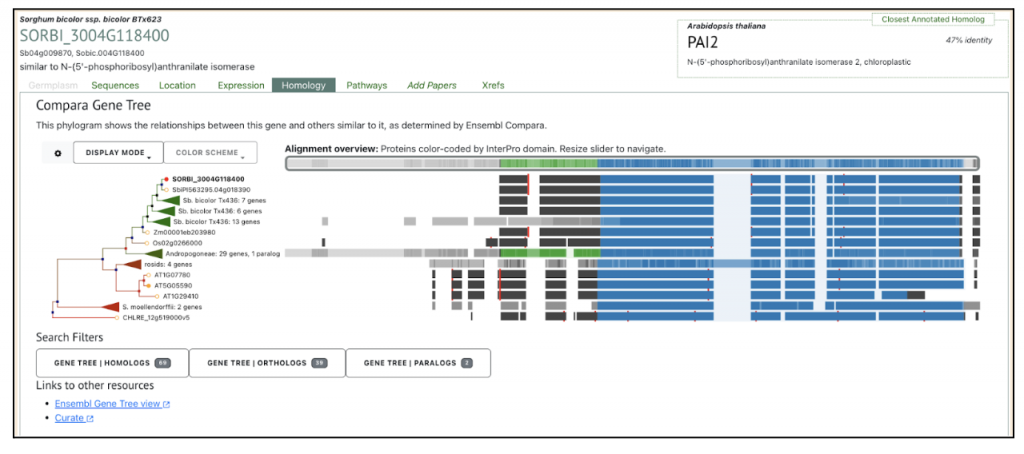
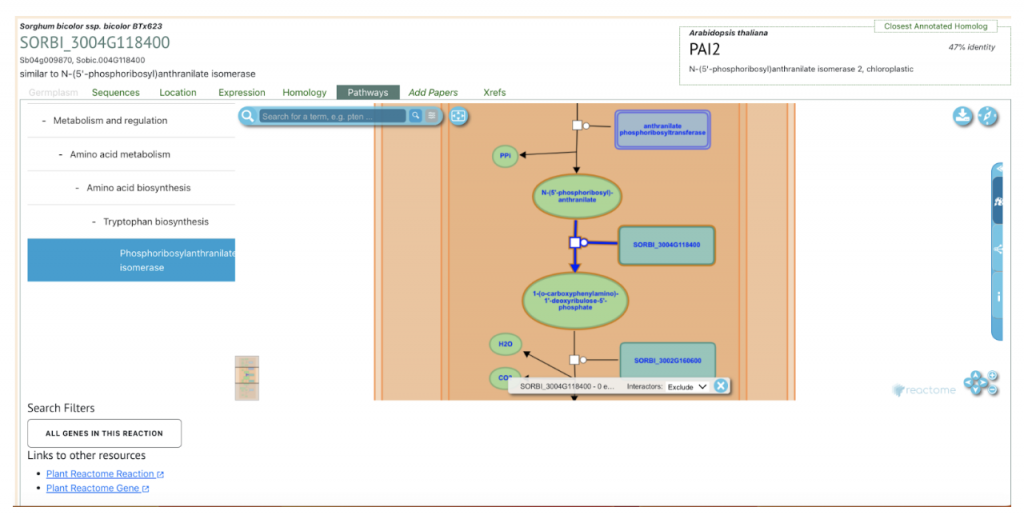
An example for Trp1 (SORBI_3004G118400) gene Trp biosynthesis pathway was analyzed in both sorghum inbred lines. The gene SORBI_3004G118400 is a key focus in the study, particularly in the context of tryptophan biosynthesis and its response to low-nitrogen (N) stress in sorghum plants. This gene is involved in the biosynthesis of tryptophan, an essential amino acid, and plays a crucial role in the plant’s response to environmental stress. Specifically, the study reports that SORBI_3004G118400 was significantly upregulated in the low-N-tolerance 398B genotype compared to the low-N-sensitive CS3541 genotype after five days of low-N treatment. This upregulation suggests that this gene may be involved in the adaptive responses of sorghum plants to low-N stress, potentially contributing to enhanced low-N tolerance and root growth. The gene’s expression pattern and its role in tryptophan biosynthesis were confirmed through RT-qPCR, which showed a high correlation coefficient (R2 = 0.93) between the RNA-seq data and the experimental results. The transcriptome analysis in the study utilized both leaves and roots from the sorghum inbred lines (398B and CS3541) under normal and low-nitrogen (N) conditions for one and five days.
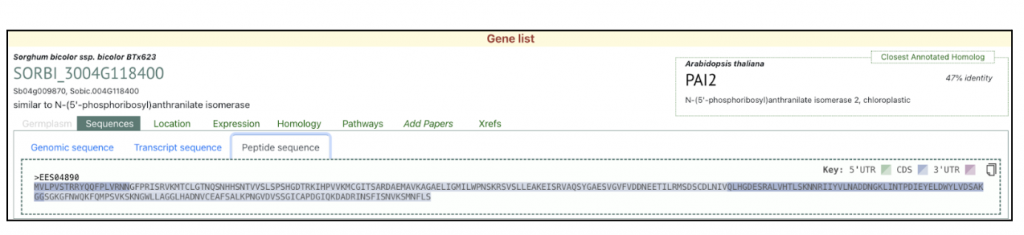
The Trp1 gene (SORBI_3004G118400) from this published study has been selected as the candidate gene to explore SorghumBase (https://sorghumbase.org/).
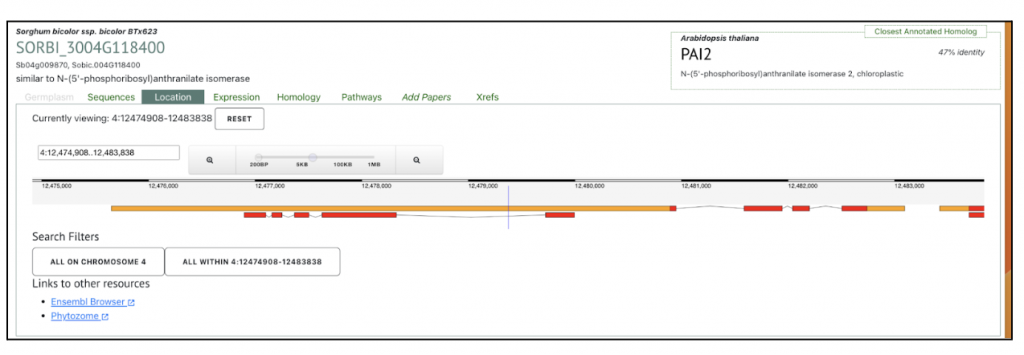
under the Location tab in the Search results in SorghumBase. It shows that the gene is located on chromosome 4; all within 4:12474908-12483838. In addition to SorghumBase, links are provided on additional resources Ensembl Browser and Phytozome to explore this gene in detail.
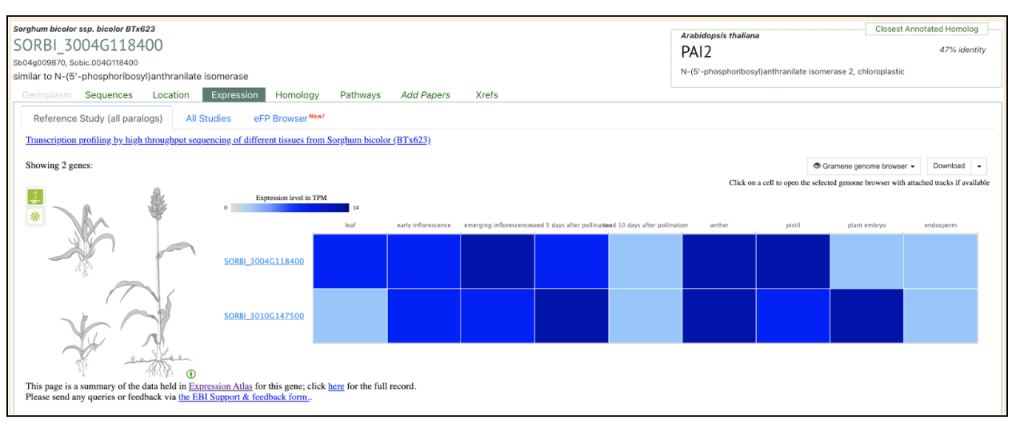
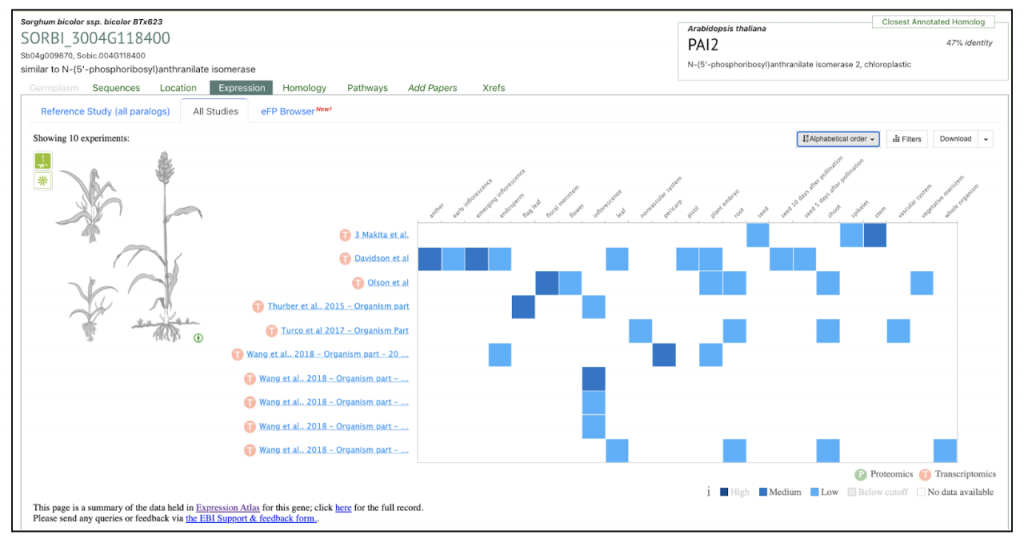
under the Expression tab (All Studies) in the search display in SorghumBase. It displays the baseline gene expression of the geneSORBI_3004G118400 in all the 10 published sorghum BTx623 datasets curated and processed by EMBL-EBI atlas in collaboration with the SorghumBase team.
Reference:
Zhao XR, Zhao DT, Zhang LY, Chang JH, Cui JH. Combining transcriptome and metabolome analysis to understand the response of sorghum to Melanaphis sacchari. BMC Plant Biol. 2024 Jun 11;24(1):529. PMID: 38862926. doi: 10.1186/s12870-024-05229-8. Read more

