Inadvertent Introgression: Pleiotropy and the Sorghum Genome
Pleiotropy, when a single gene is responsible for multiple unrelated traits, is important to identify in order to avoid unintentionally selecting for traits. Although there is little evidence for pleiotropy in maize, research suggests that the incidence may be far greater in sorghum. Identifying these traits and reducing the inadvertent introgression of non-targeted traits into elite germplasm is of great interest to breeders, biologist, and for both the grain and bioenergy industries.
Researchers at the University of Nebraska-Lincoln, Clemson University and Iowa State University assembled a set of 234 separate trait datasets for the Sorghum Association Panel (SAP) from both their own research and published studies from research groups across the country and compared genome-wide association studies with two independently generated marker sets. The researchers concluded that the existing genetic markers likely capture only 35–43% of potentially detectable loci controlling variation for traits scored in this population. In fact, when using these conventional markers through cross-GWAS comparisons, little evidence of pleiotropy was detected. The scientists determined that by increasing the marker density in the SAP samples, future studies could more than double the number of true marker-trait associations (MTAs) detected. Here, loci identified in a joint analysis of 176 trait datasets tended to fall into one of two categories, either showing associations with large (>40) number of trait datasets or small (<10) trait datasets, with these datasets often representing measures of the same trait in multiple environments or multiple distinct but highly correlated traits’, clear evidence of pleiotropic quantitative genetic loci. Both known and previously unknown pleiotropic genes were discovered, including an impact of dwarfing genes on root architecture, as well as new pleiotropic loci consistent with known trade-offs in sorghum development.
“This study demonstrates the enduring power of community association populations,” said Ravi Mural, the lead author of the study. “A single population, the SAP, has been used by so many researchers to understand the genes controlling different traits and now, by bringing all of their data together, we can start to understand the genetic basis of quantitative pleiotropy in sorghum. No one research group has the resources to create a dataset like the one we analyzed in this paper.”
SorghumBase examples:

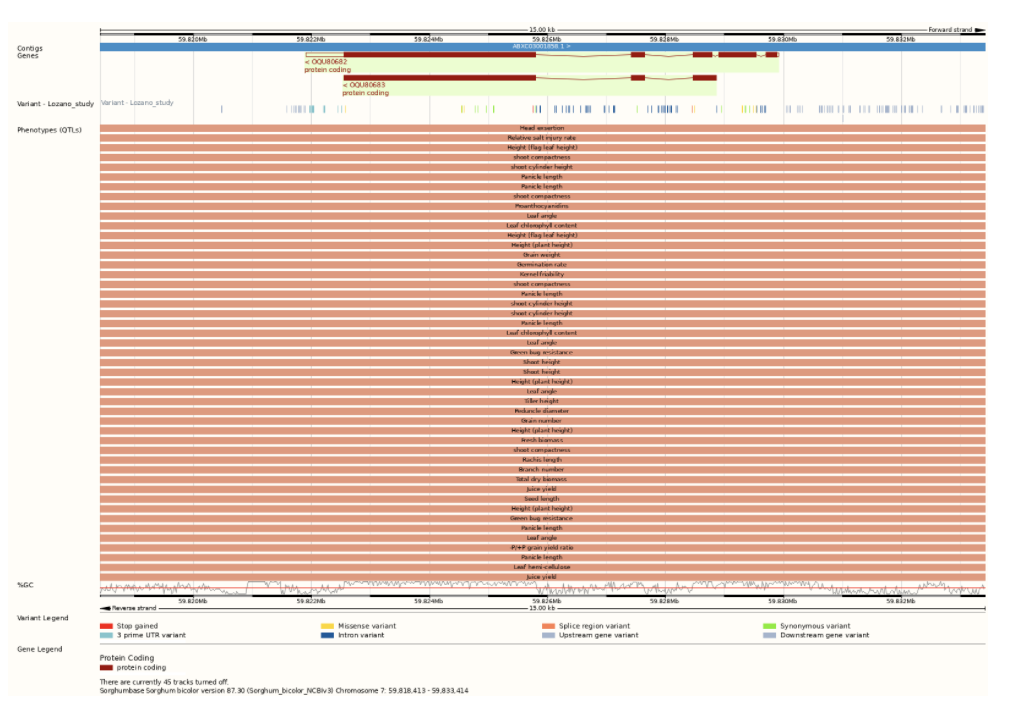
Reference
Mural, R.V., Grzybowski, M., Miao, C., Damke, A., Sapkota, S., Boyles, R.E., Salas Fernandez, M.G., Schnable, P.S., Sigmon,B., Kresovich, S., and Schnable, J.C. Meta-analysis identifies pleiotropic loci controlling phenotypic trade-offs in sorghum. Genetics, 218(3), (2021). PMID: 34100945. DOI:10.1093/genetics/iyab087. Read more
Related Project Websites:
- The Schnable Lab: https://schnablelab.org/
- University Of Nebraska-Lincoln: https://agronomy.unl.edu/schnable
- Center for Plant Science Innovation at the University of Nebraska-Lincoln: https://www.unl.edu/psi/ravi-v-mural https://www.unl.edu/psi/james-schnable




