Identification of SbGATA22 as a Negative Regulator of Dhurrin Biosynthesis in Sorghum
Cyanogenic glucosides (CNglcs) are specialized metabolites present in over 3000 higher plant species and play crucial roles in plant defense and stress mitigation. The regulation of CNglc biosynthesis, particularly in sorghum (Sorghum bicolor), remains poorly understood despite the identification of its biosynthetic pathway for dhurrin over two decades ago. Scientists from Monash University and The University of Queensland utilized yeast one-hybrid screens to identify regulatory proteins interacting with the promoter region of SbCYP79A1 (Sb01g001200; SORBI_3001G012300), a key gene in the dhurrin biosynthetic pathway. A 1204 base pair fragment of the SbCYP79A1 promoter was cloned and used as bait in yeast, which was subsequently transformed with cDNA libraries from different sorghum developmental stages. This approach led to the identification of SbGATA22, an LLM domain B-GATA transcription factor that binds to putative GATA motifs within the SbCYP79A1 promoter region. Further analysis showed SbGATA22 localizes to the nucleus and may act as a negative regulator of SbCYP79A1 expression, suggesting a role in the molecular regulation of dhurrin biosynthesis in sorghum.
The study also examined the expression of SbGATA22 in correlation with SbCYP79A1 and dhurrin concentration over 14 days of sorghum development and under nitrogen application, a known factor affecting dhurrin levels. Results indicated that SbGATA22 expression is inversely correlated with SbCYP79A1 during development and nitrogen treatment, supporting its potential role as a repressor. This research provides initial insights into the transcriptional regulation of dhurrin biosynthesis and highlights the complexity of CNglc regulation in response to environmental factors. These findings open avenues for more precise manipulation of CNglc levels in sorghum, aiming to mitigate the risks associated with cyanide toxicity in forage while preserving plant stress tolerance and growth.
SorghumBase examples:
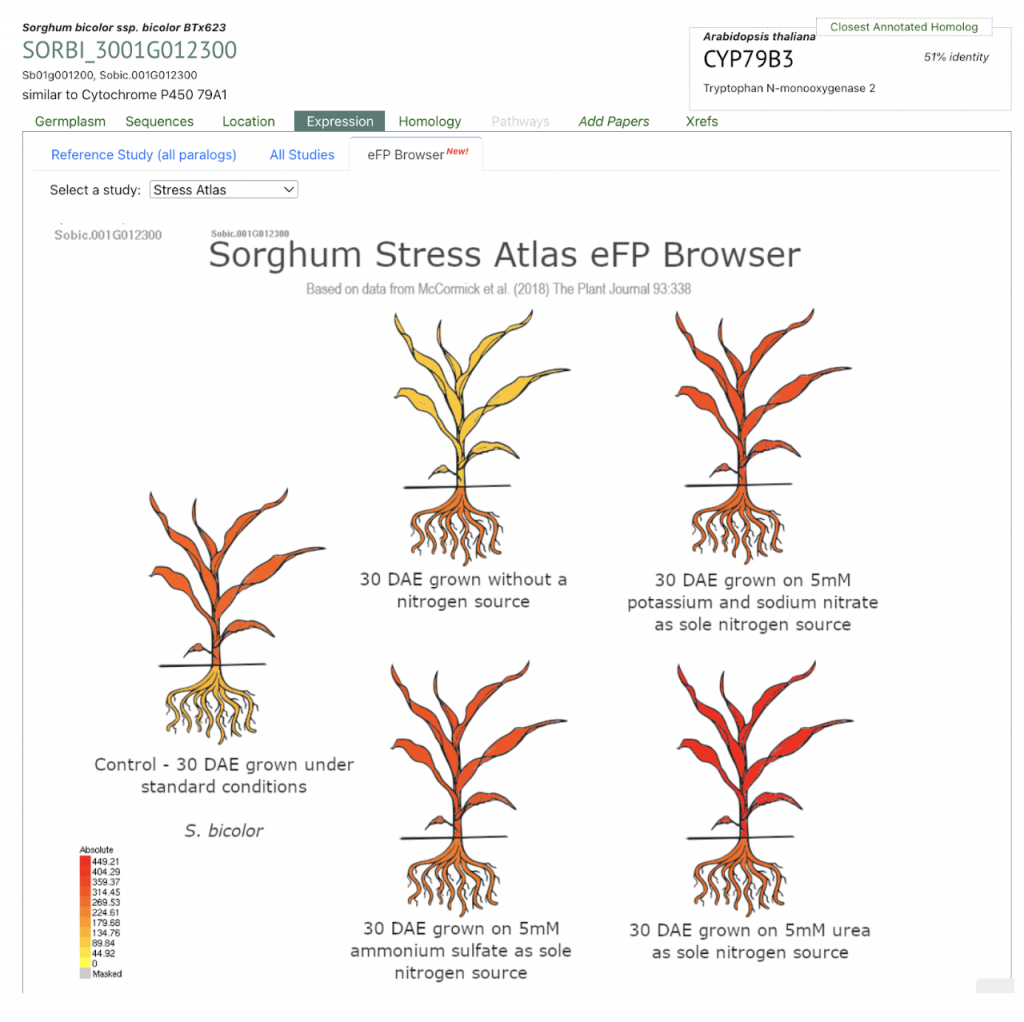
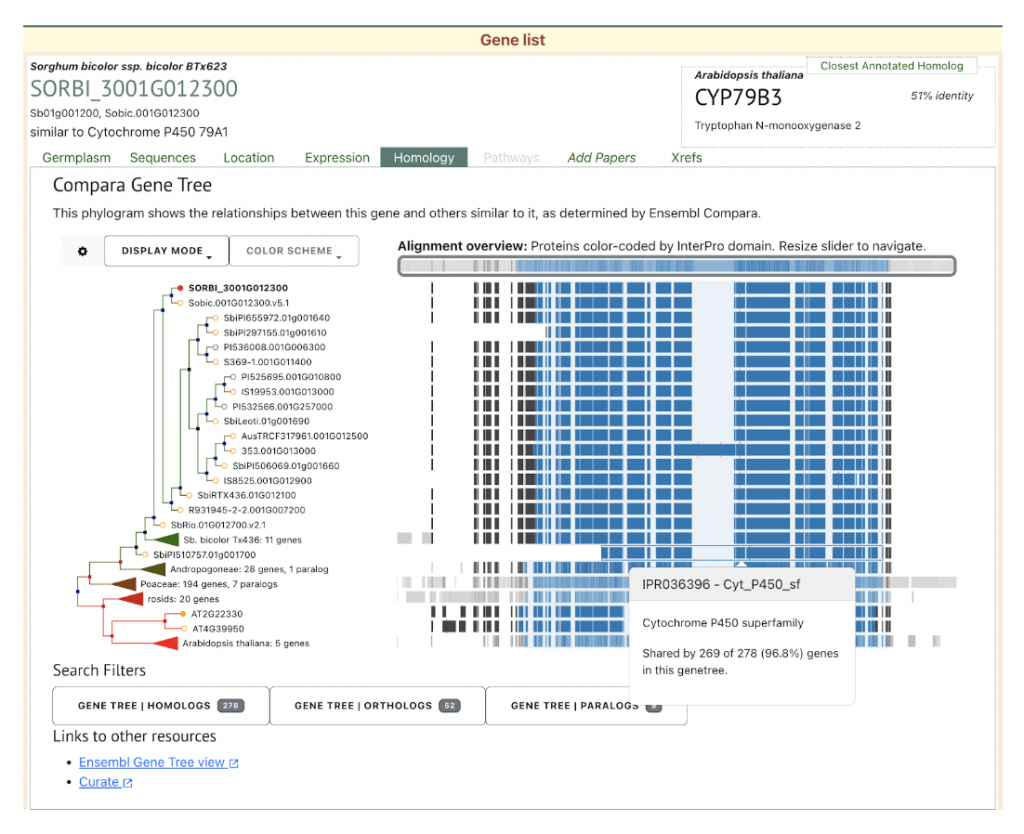

Reference:
Rosati VC, Quinn AA, Gleadow RM, Blomstedt CK. The Putative GATA Transcription Factor SbGATA22 as a Novel Regulator of Dhurrin Biosynthesis. Life (Basel). 2024 Apr 3;14(4):470. PMID: 38672741. doi: 10.3390/life14040470. Read more
Related Project Websites:
Dr. Roslyn Gleadow’s page at Monash University: https://www.monash.edu/science/schools/biological-sciences/staff/ros-gleadow




