Identification of Marker-Trait Associations for Sorghum Aphid Resistance
Sorghum (Sorghum bicolor (L). Moench), used for food, livestock feed and biofuel, is the fifth most important cereal crop globally. The largest producer and exporter of grain sorghum is the United States. Fields throughout the United States have been invaded by the sorghum aphid (SA), Melanaphis sorghi (Theobald) causing decreased grain yield, reduced seed weight, lower grain quality, increased lodging, aborted panicle emergence and plant death. Practices such as the use of pesticides and natural predators have helped combat the issue; however, plant resistance also plays an important role in reducing yield loss and damage to crops. Many sorghum inbred lines that are resistant to SA have been identified while, conversely, few of the genes behind this trait have been identified. The main previously discovered genes associated with resistance to SA are the RMES1 locus and Quantitative Trait Loci identified on chromosome 6. Expanding knowledge of sorghum resistant lines is of great interest to scientists, breeders and growers.
In an effort to identify additional genes involved in resistance to the sorghum aphid, researchers from Fort Valley State University in Georgia, the USDA-ARS, and the University of Georgia used the Sorghum Association Panel (SAP) and some additional lines, to pinpoint new genomic regions connected to resistance to SA. The researchers gathered phenotypic data in both 2019 and 2020 in Tifton, GA, using visual inspection and, in 2020 additionally used drone-based high-throughput phenotyping. They then used Genotyping-by-Sequencing data to identify Marker-Trait Associations through a Genome-Wide Association Study. Seventy markers for six aphid resistance-related traits were identified, sixty of which were at unique locations. The researchers consistently found that the sorghum plants that had SA resistance traits had co-locating SNPs on chromosomes 2, 3, 5, 8 and 10 across different traits. These regions encode genes including leucine-rich repeats (LRR), Avr proteins, lipoxygenases (LOXs), calmodulins (CAM) dependent protein kinase, WRKY transcription factors, flavonoid biosynthesis genes, and 12-oxo-phytodienoic acid reductase. Several of these markers could be used in genomic selection to produce improved SA resistant cultivars, though confirmation of the results in future studies through functional validation and positional cloning is necessary.
This work was impossible without having vast amounts of genetic and genomic resources in the sorghum research community. SorghumBase platform was vital in providing the sorghum genome database for identification of relevant genes for this manuscript. This study identified three new sources of resistant lines to sorghum aphid and genomic regions, which provide support in dissecting the causative genes involved in sorghum aphid resistance. – Punnuri
These aphids are a real problem for anyone growing sorghum. I’ve seen them completely destroy a lot of varieties, and as you can imagine, trying to count how many aphids are on the underside of leaves for a few hundred varieties can get old fast—especially in Georgia summers. Being able to use remote sensing to get even a first approximation of resistance would help things go much faster and more efficiently to get new resistance loci identified and deployed to farmers’ fields. – Wallace
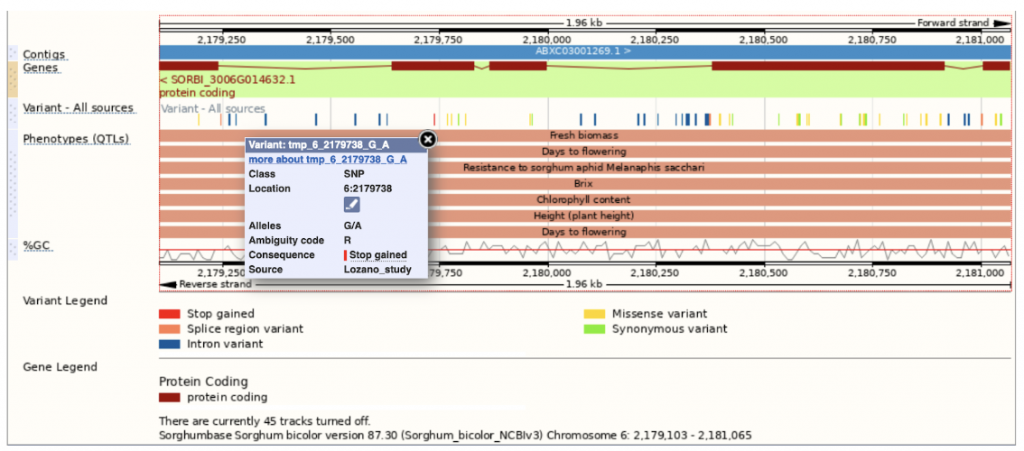
SorghumBase example:
Reference:
Punnuri SM, Ayele AG, Harris-Shultz KR, Knoll JE, Coffin AW, Tadesse HK, Armstrong JS, Wiggins TK, Li H, Sattler S, Wallace JG. Genome-wide association mapping of resistance to the sorghum aphid in Sorghum bicolor. Genomics. 2022 Jun 15;114(4):110408. PMID: 35716823. DOI: 10.1016/j.ygeno.2022.110408. Read more
Related Project Websites:
The Wallace Lab: http://wallacelab.uga.edu/people.html
Dr. Punnuri at Fort Valley State University: https://ag.fvsu.edu/members/profile/view/63


