Diversity in the Sorghum Pan-Genome Could Contribute to Crop Improvement
Sorghum, a staple food for people in Africa and Asia, is drought and stress tolerant. It is a promising crop given the changing global climate, its use as a food source, feed source for livestock, and biofuel, and the increasing demands of an ever increasing population. Understanding Sorghum’s genetic diversity in both its domesticated and wild strains is important for crop improvement and gene discovery and is of interest to both biologists and breeders.
To enable the investigation of sorghum diversity, researchers at the University of Queensland in Queensland, Australia, the Chinese Academy of Sciences in Beijing, China, and BGI Genomics in Shenzhen, China analyzed a sorghum pan-genome within the sorghum primary gene pool. They assembled 13 genomes from both domesticated and wild sorghum varieties and with 3 other published genomes generated a pan-genome of 44,079 gene families with 222.6 Mb of new sequence identified. 64% of gene families showed presence/ absence variation, which is substantial. Transposable elements and differing recombination rates influenced the distribution of these variants across the genomes. The identified presence/absence variants that were selected for during domestication had phenotypic consequences that could be associated with crop improvement.
“We are amazed to see the substantial gene content variations among sorghum lines, which highlights the great potential of sorghum and could help better understand heterosis in sorghum.” Dr Yongfu Tao explained.
“Sorghum is a highly diverse species, having access to information on which sorghum lines contain which genes is a paradigm-shift for future sorghum research and improvement activities” Dr. Emma Mace said.
“Our approach was practical, with the project designed to have applications in breeding that help drive gains in productivity for the grains industry,” Professor David Jordan said.
SorghumBase example:
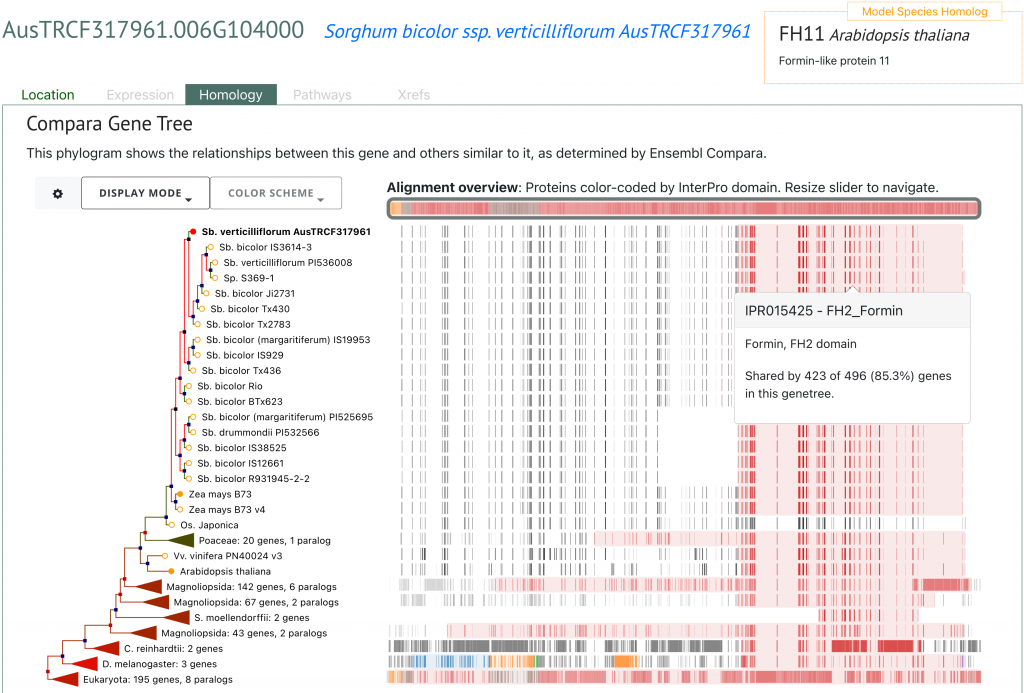
Reference
Tao, Y., Luo, H., Xu, J., Cruickshank, A., Zhao, X., Teng, F., Hathorn, A., Wu, X., Liu, Y., Shatte, T., Jordan, D., Jing, H., and Mace, E. Extensive variation within the pan-genome of cultivated and wild sorghum. Nat. Plants 7, 766–773 (2021). https://doi.org/10.1038/s41477-021-00925-x. Read more
Related Project Websites
-

Image 1: Professor David Jordan (left), Dr. Emma Mace (middle) and Dr. Yongfu Tao (right) in sorghum field. Credit from Cassie Martinez.

